Base excision repair of oxidative DNA damage: from mechanism to disease
- PMID: 28199214
- PMCID: PMC5567671
- DOI: 10.2741/4555
Base excision repair of oxidative DNA damage: from mechanism to disease
Abstract
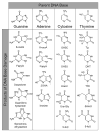
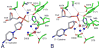
Reactive oxygen species continuously assault the structure of DNA resulting in oxidation and fragmentation of the nucleobases. Both oxidative DNA damage itself and its repair mediate the progression of many prevalent human maladies. The major pathway tasked with removal of oxidative DNA damage, and hence maintaining genomic integrity, is base excision repair (BER). The aphorism that structure often dictates function has proven true, as numerous recent structural biology studies have aided in clarifying the molecular mechanisms used by key BER enzymes during the repair of damaged DNA. This review focuses on the mechanistic details of the individual BER enzymes and the association of these enzymes during the development and progression of human diseases, including cancer and neurological diseases. Expanding on these structural and biochemical studies to further clarify still elusive BER mechanisms, and focusing our efforts toward gaining an improved appreciation of how these enzymes form co-complexes to facilitate DNA repair is a crucial next step toward understanding how BER contributes to human maladies and how it can be manipulated to alter patient outcomes.
Figures

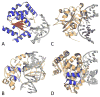
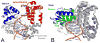

References
-
- Jovanovic SV, Simic MG. One-Electron Redox Potentials of Purines and Pyrimidines. Journal of Physical Chemistry. 1986;90(5):974–978. doi: 10.1021/j100277a053. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

