Host Genotype and Gut Microbiome Modulate Insulin Secretion and Diet-Induced Metabolic Phenotypes
- PMID: 28199845
- PMCID: PMC5325228
- DOI: 10.1016/j.celrep.2017.01.062
Host Genotype and Gut Microbiome Modulate Insulin Secretion and Diet-Induced Metabolic Phenotypes
Abstract
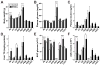
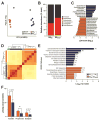
Genetic variation drives phenotypic diversity and influences the predisposition to metabolic disease. Here, we characterize the metabolic phenotypes of eight genetically distinct inbred mouse strains in response to a high-fat/high-sucrose diet. We found significant variation in diabetes-related phenotypes and gut microbiota composition among the different mouse strains in response to the dietary challenge and identified taxa associated with these traits. Follow-up microbiota transplant experiments showed that altering the composition of the gut microbiota modifies strain-specific susceptibility to diet-induced metabolic disease. Animals harboring microbial communities with enhanced capacity for processing dietary sugars and for generating hydrophobic bile acids showed increased susceptibility to metabolic disease. Notably, differences in glucose-stimulated insulin secretion between different mouse strains were partially recapitulated via gut microbiota transfer. Our results suggest that the gut microbiome contributes to the genetic and phenotypic diversity observed among mouse strains and provide a link between the gut microbiome and insulin secretion.
Keywords: gut microbiome; insulin secretion; metabolic disease; pancreatic islets.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

