Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies
- PMID: 28200038
- PMCID: PMC5834090
- DOI: 10.1093/annonc/mdw683
Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies
Abstract
Background: A more accurate prognosis for non-small-cell lung cancer (NSCLC) patients could aid in the identification of patients at high risk for recurrence. Many NSCLC mRNA expression signatures claiming to be prognostic have been reported in the literature. The goal of this study was to identify the most promising mRNA prognostic signatures in NSCLC for further prospective clinical validation.
Experimental design: We carried out a systematic review and meta-analysis of published mRNA prognostic signatures for resected NSCLC. The prognostic performance of each signature was evaluated via a meta-analysis of 1927 early stage NSCLC patients collected from 15 studies using three evaluation metrics (hazard ratios, concordance scores, and time-dependent receiver-operating characteristic curves). The performance of each signature was then evaluated against 100 random signatures. The prognostic power independent of clinical risk factors was assessed by multivariate Cox models.
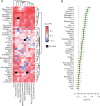
Results: Through a literature search, we identified 42 lung cancer prognostic signatures derived from genome-wide expression profiling analysis. Based on meta-analysis, 25 signatures were prognostic for survival after adjusting for clinical risk factors and 18 signatures carried out significantly better than random signatures. When analyzing histology types separately, 17 signatures and 8 signatures are prognostic for adenocarcinoma and squamous cell lung cancer, respectively. Despite little overlap among published gene signatures, the top-performing signatures are highly concordant in predicted patient outcomes.
Conclusions: Based on this large-scale meta-analysis, we identified a set of mRNA expression prognostic signatures appropriate for further validation in prospective clinical studies.
Keywords: meta-analysis; non-small-cell lung cancer; prognostic gene signatures.
© The Author 2017. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- National Comprehensive Cancer Network I. In NCCN Clinical Practice Guidelines in Oncology. Non-small Cell Lung Cancer 2016 (Version 4.2016).
-
- Tanoue LT. Staging of non-small cell lung cancer. Semin Respir Crit Care Med 2008; 29: 248–260. - PubMed
-
- Subramanian J, Simon R.. What should physicians look for in evaluating prognostic gene-expression signatures? Nat Rev Clin Oncol 2010; 7: 327–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

