Adaptive MCMC in Bayesian phylogenetics: an application to analyzing partitioned data in BEAST
- PMID: 28200071
- PMCID: PMC6044345
- DOI: 10.1093/bioinformatics/btx088
Adaptive MCMC in Bayesian phylogenetics: an application to analyzing partitioned data in BEAST
Abstract
Motivation: Advances in sequencing technology continue to deliver increasingly large molecular sequence datasets that are often heavily partitioned in order to accurately model the underlying evolutionary processes. In phylogenetic analyses, partitioning strategies involve estimating conditionally independent models of molecular evolution for different genes and different positions within those genes, requiring a large number of evolutionary parameters that have to be estimated, leading to an increased computational burden for such analyses. The past two decades have also seen the rise of multi-core processors, both in the central processing unit (CPU) and Graphics processing unit processor markets, enabling massively parallel computations that are not yet fully exploited by many software packages for multipartite analyses.
Results: We here propose a Markov chain Monte Carlo (MCMC) approach using an adaptive multivariate transition kernel to estimate in parallel a large number of parameters, split across partitioned data, by exploiting multi-core processing. Across several real-world examples, we demonstrate that our approach enables the estimation of these multipartite parameters more efficiently than standard approaches that typically use a mixture of univariate transition kernels. In one case, when estimating the relative rate parameter of the non-coding partition in a heterochronous dataset, MCMC integration efficiency improves by > 14-fold.
Availability and implementation: Our implementation is part of the BEAST code base, a widely used open source software package to perform Bayesian phylogenetic inference.
Contact: guy.baele@kuleuven.be.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com
Figures
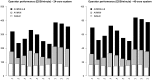
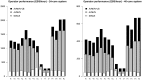
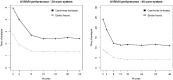
References
-
- Baele G., Lemey P. (2013) Bayesian evolutionary model testing in the phylogenomics era: matching model complexity with computational efficiency. Bioinformatics, 29, 1970–1979. - PubMed
-
- Ferreira M.A.R., Suchard M.A. (2008) Bayesian anaylsis of elasped times in continuous-time Markov chains. Canadian Journal of Statistics, 26, 355–368.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

