Patterns and processes of Mycobacterium bovis evolution revealed by phylogenomic analyses
- PMID: 28201585
- PMCID: PMC5381553
- DOI: 10.1093/gbe/evx022
Patterns and processes of Mycobacterium bovis evolution revealed by phylogenomic analyses
Abstract
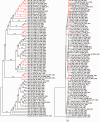
Mycobacterium bovis is an important animal pathogen worldwide that parasitizes wild and domesticated vertebrate livestock as well as humans. A comparison of the five M. bovis complete genomes from the United Kingdom, South Korea, Brazil, and the United States revealed four novel large-scale structural variations of at least 2,000 bp. A comparative phylogenomic study including 2,483 core genes of 38 taxa from eight countries showed conflicting phylogenetic signal among sites. By minimizing this effect, we obtained a tree that better agrees with sampling locality. Results supported a relatively basal position of African strains (all isolated from Homo sapiens), confirming that Africa was an important region for early diversification and that humans were one of the earliest hosts. Selection analyses revealed that functional categories such as “Lipid transport and metabolism,” “Cell cycle control, cell division, chromosome partitioning” and “Cell motility” were significant for the evolution of the group, besides other categories previously described, showing importance of genes associated with virulence and cholesterol metabolism in the evolution of M. bovis. PE/PPE genes, many of which are known to be associated with virulence, were major targets for large-scale polymorphisms, homologous recombination, and positive selection, evincing for the first time a plethora of evolutionary forces possibly contributing to differential adaptability in M. bovis. By assuming different priors, US strains originated and started to diversify around 150–5,210 ya. By further analyzing the largest set of US genomes to date (76 in total), obtained from 14 host species, we detected that hosts were not clustered in clades (except for a few cases), with some faster-evolving strains being detected, suggesting fast and ongoing reinfections across host species, and therefore, the possibility of new bovine tuberculosis outbreaks.
Figures



References
-
- Abdallah A, et al. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 62:667–679. - PubMed
-
- Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. 2012. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie. 94:110–116. - PubMed
-
- Allen AR, et al. 2013. The phylogeny and population structure of Mycobacterium bovis in the British Isles. Infect Genet Evol. 20:8–15. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

