Comprehensive molecular characterization of multifocal glioblastoma proves its monoclonal origin and reveals novel insights into clonal evolution and heterogeneity of glioblastomas
- PMID: 28201779
- PMCID: PMC5464316
- DOI: 10.1093/neuonc/now231
Comprehensive molecular characterization of multifocal glioblastoma proves its monoclonal origin and reveals novel insights into clonal evolution and heterogeneity of glioblastomas
Abstract
Background: The evolution of primary glioblastoma (GBM) is poorly understood. Multifocal GBM (ie, multiple synchronous lesions in one patient) could elucidate GBM development.
Methods: We present the first comprehensive study of 12 GBM foci from 6 patients using array-CGH, spectral karyotyping, gene expression arrays, and next-generation sequencing.
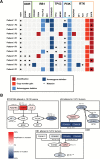
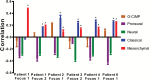
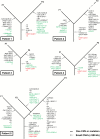
Results: Multifocal GBMs genetically resemble primary GBMs. Comparing foci from the same patient proved their monoclonal origin. All tumors harbored alterations in the 3 GBM core pathways: RTK/PI3K, p53, and RB regulatory pathways with aberrations of EGFR and CDKN2A/B in all (100%) patients. This unexpected high frequency reflects a distinct genetic signature of multifocal GBMs and might account for their highly malignant and invasive phenotype. Surprisingly, the types of mutations in these genes/pathways were different in tumor foci from the same patients. For example, we found distinct mutations/aberrations in PTEN, TP53, EGFR, and CDKN2A/B, which therefore must have occurred independently and late during tumor development. We also identified chromothripsis as a late event and in tumors with wild-type TP53. Only 2 events were found to be early in all patients: single copy loss of PTEN and TERT promoter point mutations.
Conclusions: Multifocal GBMs develop through parallel genetic evolution. The high frequency of alterations in 3 main pathways suggests that these are essential steps in GBM evolution; however, their late occurrence indicates that they are not founder events but rather subclonal drivers. This might account for the marked genetic heterogeneity seen in primary GBM and therefore has important implications for GBM therapy.
Keywords: monoclonal origin; multifocal glioblastoma; tumor evolution; tumor genetics; tumor heterogeneity.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures


Comment in
-
Discordance of IDH mutational status between lesions in an adult patient with multifocal glioma.Neuro Oncol. 2018 Jul 5;20(8):1142-1143. doi: 10.1093/neuonc/noy080. Neuro Oncol. 2018. PMID: 29868765 Free PMC article. No abstract available.
References
-
- Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. - PubMed
-
- Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

