Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism
- PMID: 28201981
- PMCID: PMC5310080
- DOI: 10.1186/s12863-017-0481-y
Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism
Abstract
Background: Trichoderma spp. can establish beneficial interactions with plants by promoting plant growth and defense systems, as well as, antagonizing fungal phytopathogens in mycoparasitic interactions. Such interactions depend on signal exchange between both participants and can be mediated by effector proteins that alter the host cell structure and function, allowing the establishment of the relationship. The main purpose of this work was to identify, using computational methods, candidates of effector proteins from T. virens, T. atroviride and T. reesei, validate the expression of some of the genes during a beneficial interaction and mycoparasitism and to define the biological function for one of them.
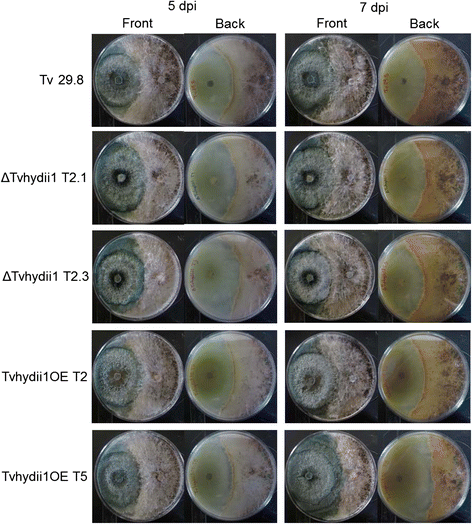
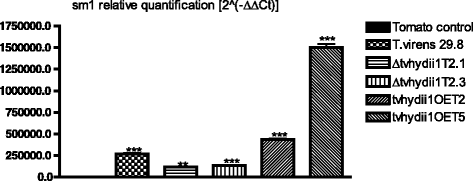
Results: We defined a catalogue of putative effector proteins from T. virens, T. atroviride and T. reesei. We further validated the expression of 16 genes encoding putative effector proteins from T. virens and T. atroviride during the interaction with the plant Arabidopsis thaliana, and with two anastomosis groups of the phytopathogenic fungus Rhizoctonia solani. We found genes which transcript levels are modified in response to the presence of both plant fungi, as well as genes that respond only to either a plant or a fungal host. Further, we show that overexpression of the gene tvhydii1, a Class II hydrophobin family member, enhances the antagonistic activity of T. virens against R. solani AG2. Further, deletion of tvhydii1 results in reduced colonization of plant roots, while its overexpression increases it.
Conclusions: Our results show that Trichoderma is able to respond in different ways to the presence of a plant or a fungal host, and it can even distinguish between different strains of fungi of a given species. The putative effector proteins identified here may play roles in preventing perception of the fungus by its hosts, favoring host colonization or protecting it from the host's defense response. Finally, the novel effector protein TVHYDII1 plays a role in plant root colonization by T, virens, and participates in its antagonistic activity against R. solani.
Keywords: Effector; Hydrophobin; Mycoparasitism; Plant-fungus interaction; Trichoderma.
Figures






Similar articles
-
IPA-1 a Putative Chromatin Remodeler/Helicase-Related Protein of Trichoderma virens Plays Important Roles in Antibiosis Against Rhizoctonia solani and Induction of Arabidopsis Systemic Disease Resistance.Mol Plant Microbe Interact. 2020 Jun;33(6):808-824. doi: 10.1094/MPMI-04-19-0092-R. Epub 2020 Apr 16. Mol Plant Microbe Interact. 2020. PMID: 32101077
-
Argonaute and Dicer are essential for communication between Trichoderma atroviride and fungal hosts during mycoparasitism.Microbiol Spectr. 2024 Apr 2;12(4):e0316523. doi: 10.1128/spectrum.03165-23. Epub 2024 Mar 5. Microbiol Spectr. 2024. PMID: 38441469 Free PMC article.
-
Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism.BMC Genomics. 2013 Feb 22;14:121. doi: 10.1186/1471-2164-14-121. BMC Genomics. 2013. PMID: 23432824 Free PMC article.
-
Plant-beneficial effects of Trichoderma and of its genes.Microbiology (Reading). 2012 Jan;158(Pt 1):17-25. doi: 10.1099/mic.0.052274-0. Epub 2011 Oct 13. Microbiology (Reading). 2012. PMID: 21998166 Review.
-
Self versus non-self: fungal cell wall degradation in Trichoderma.Microbiology (Reading). 2012 Jan;158(Pt 1):26-34. doi: 10.1099/mic.0.052613-0. Epub 2011 Aug 26. Microbiology (Reading). 2012. PMID: 21873410 Review.
Cited by
-
Hydrophobin HFBII-4 from Trichoderma asperellum induces antifungal resistance in poplar.Braz J Microbiol. 2019 Jul;50(3):603-612. doi: 10.1007/s42770-019-00083-5. Epub 2019 Apr 13. Braz J Microbiol. 2019. PMID: 30982213 Free PMC article.
-
Trichoderma as a Model to Study Effector-Like Molecules.Front Microbiol. 2019 May 15;10:1030. doi: 10.3389/fmicb.2019.01030. eCollection 2019. Front Microbiol. 2019. PMID: 31156578 Free PMC article. Review.
-
Characterization of Three Fusarium graminearum Effectors and Their Roles During Fusarium Head Blight.Front Plant Sci. 2020 Nov 30;11:579553. doi: 10.3389/fpls.2020.579553. eCollection 2020. Front Plant Sci. 2020. PMID: 33329641 Free PMC article.
-
The Biocontrol and Growth-Promoting Potential of Penicillium spp. and Trichoderma spp. in Sustainable Agriculture.Plants (Basel). 2025 Jun 30;14(13):2007. doi: 10.3390/plants14132007. Plants (Basel). 2025. PMID: 40648017 Free PMC article.
-
Biocontrol Agents: Toolbox for the Screening of Weapons against Mycotoxigenic Fusarium.J Fungi (Basel). 2021 Jun 3;7(6):446. doi: 10.3390/jof7060446. J Fungi (Basel). 2021. PMID: 34205071 Free PMC article.
References
-
- Saloheimo M, Wang H, Valkonen M, Vasara T, Huuskonen A, Riikonen M, et al. Characterization of Secretory Genes ypt1 / yptA and nsf1 / nsfA from two filamentous fungi: induction of secretory pathway genes of Trichoderma reesei under secretion stress conditions. Appl Environ Microbiol. 2004;70:459–467. doi: 10.1128/AEM.70.1.459-467.2004. - DOI - PMC - PubMed
-
- Benítez T, Rincón AM, Limón MC, Codón AC. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 2004;7:249–260. - PubMed
-
- Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, López-Bucio J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 2009;149:1579–1592. doi: 10.1104/pp.108.130369. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

