Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing
- PMID: 28201982
- PMCID: PMC5312572
- DOI: 10.1186/s12864-017-3532-x
Transcriptome profile of rat genes in injured spinal cord at different stages by RNA-sequencing
Abstract
Background: Spinal cord injury (SCI) results in fatal damage and currently has no effective treatment. The pathological mechanisms of SCI remain unclear. In this study, genome-wide transcriptional profiling of spinal cord samples from injured rats at different time points after SCI was performed by RNA-Sequencing (RNA-Seq). The transcriptomes were systematically characterized to identify the critical genes and pathways that are involved in SCI pathology.
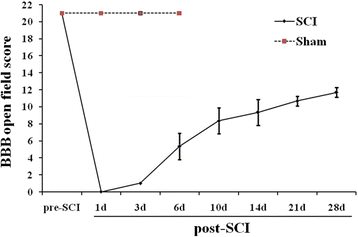
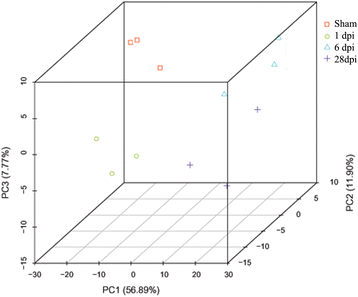
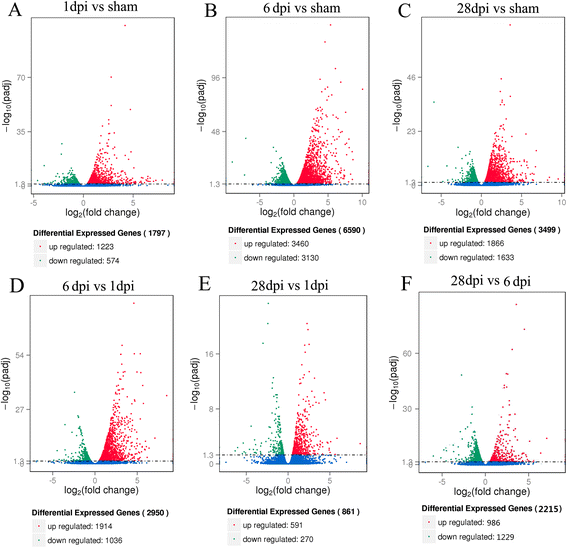
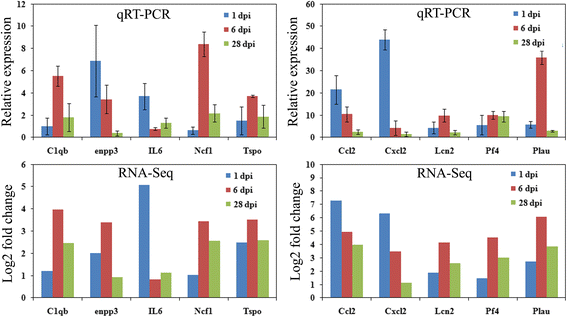
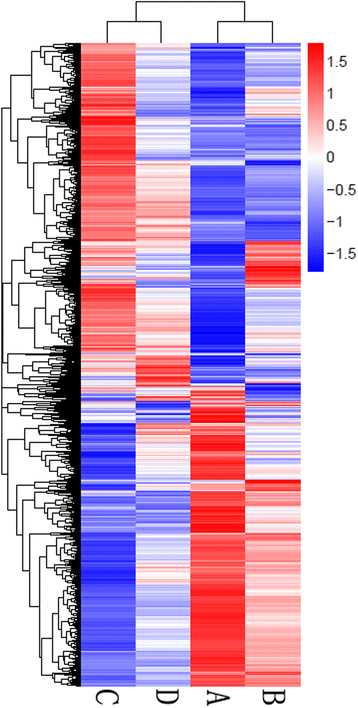
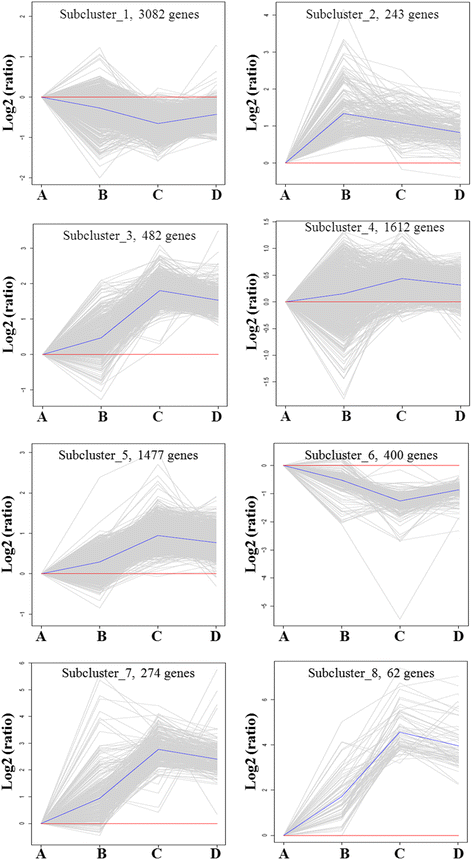
Results: RNA-Seq results were obtained from total RNA harvested from the spinal cords of sham control rats and rats in the acute, subacute, and chronic phases of SCI (1 day, 6 days and 28 days after injury, respectively; n = 3 in every group). Compared with the sham-control group, the number of differentially expressed genes was 1797 in the acute phase (1223 upregulated and 574 downregulated), 6590 in the subacute phase (3460 upregulated and 3130 downregulated), and 3499 in the chronic phase (1866 upregulated and 1633 downregulated), with an adjusted P-value <0.05 by DESeq. Gene ontology (GO) enrichment analysis showed that differentially expressed genes were most enriched in immune response, MHC protein complex, antigen processing and presentation, translation-related genes, structural constituent of ribosome, ion gated channel activity, small GTPase mediated signal transduction and cytokine and/or chemokine activity. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the most enriched pathways included ribosome, antigen processing and presentation, retrograde endocannabinoid signaling, axon guidance, dopaminergic synapses, glutamatergic synapses, GABAergic synapses, TNF, HIF-1, Toll-like receptor, NF-kappa B, NOD-like receptor, cAMP, calcium, oxytocin, Rap1, B cell receptor and chemokine signaling pathway.
Conclusions: This study has not only characterized changes in global gene expression through various stages of SCI progression in rats, but has also systematically identified the critical genes and signaling pathways in SCI pathology. These results will expand our understanding of the complex molecular mechanisms involved in SCI and provide a foundation for future studies of spinal cord tissue damage and repair. The sequence data from this study have been deposited into Sequence Read Archive ( http://www.ncbi.nlm.nih.gov/sra ; accession number PRJNA318311).
Keywords: GO enrichment; Pathway analysis; RNA-Seq; Spinal cord injury; Sprague-Dawley rats (RRID:RGD_70508).
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

