Plasmid diversity and phylogenetic consistency in the Lyme disease agent Borrelia burgdorferi
- PMID: 28201991
- PMCID: PMC5310021
- DOI: 10.1186/s12864-017-3553-5
Plasmid diversity and phylogenetic consistency in the Lyme disease agent Borrelia burgdorferi
Abstract
Background: Bacteria from the genus Borrelia are known to harbor numerous linear and circular plasmids. We report here a comparative analysis of the nucleotide sequences of 236 plasmids present in fourteen independent isolates of the Lyme disease agent B. burgdorferi.
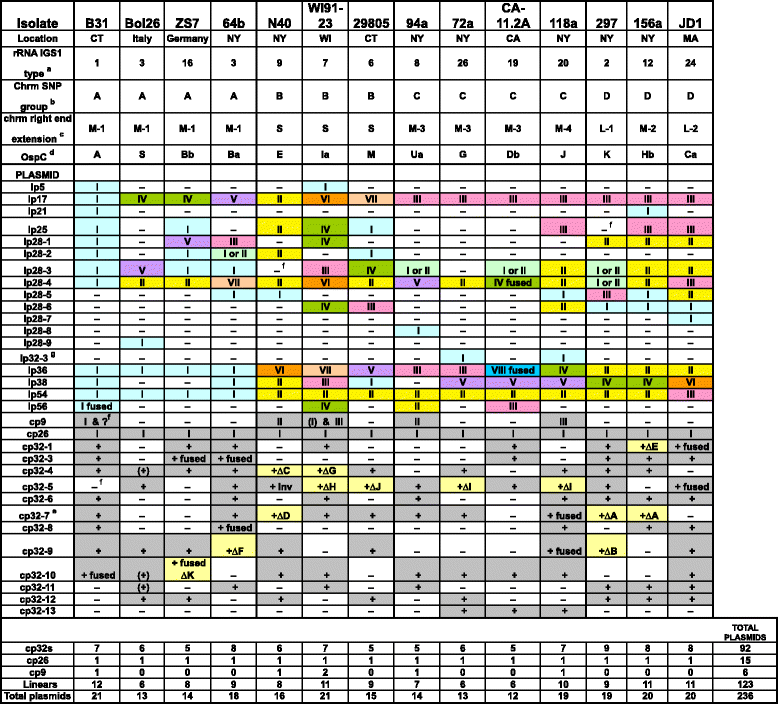
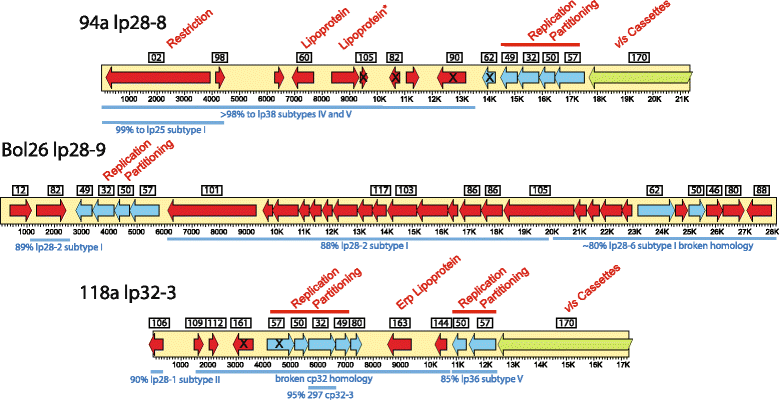
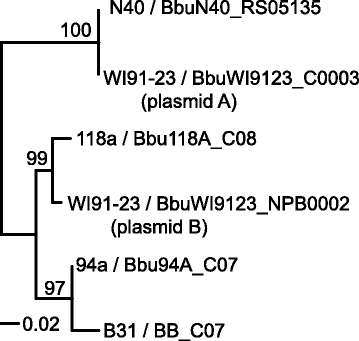
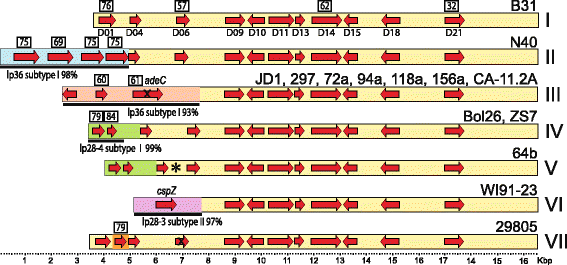
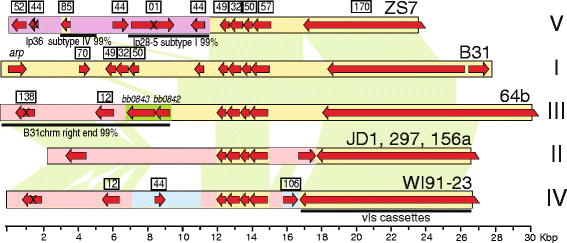
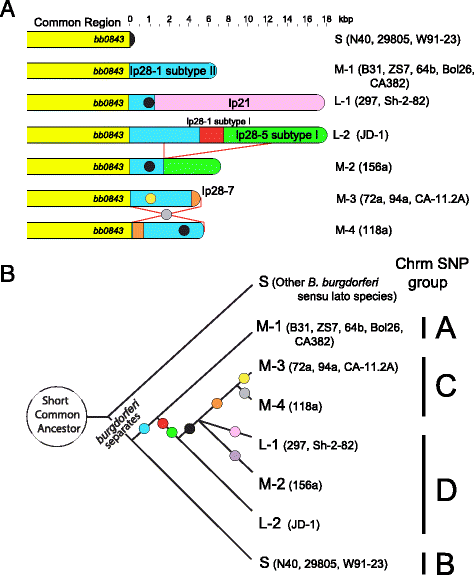
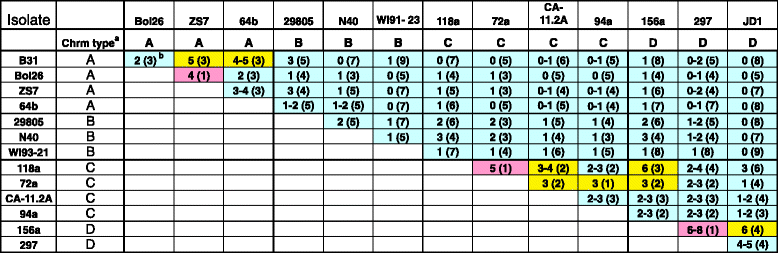
Results: We have sequenced the genomes of 14 B. burgdorferi sensu stricto isolates that carry a total of 236 plasmids. These individual isolates carry between seven and 23 plasmids. Their chromosomes, the cp26 and cp32 circular plasmids, as well as the lp54 linear plasmid, are quite evolutionarily stable; however, the remaining plasmids have undergone numerous non-homologous and often duplicative recombination events. We identify 32 different putative plasmid compatibility types among the 236 plasmids, of which 15 are (usually) circular and 17 are linear. Because of past rearrangements, any given gene, even though it might be universally present in these isolates, is often found on different linear plasmid compatibility types in different isolates. For example, the arp gene and the vls cassette region are present on plasmids of four and five different compatibility types, respectively, in different isolates. A majority of the plasmid types have more than one organizationally different subtype, and the number of such variants ranges from one to eight among the 18 linear plasmid types. In spite of this substantial organizational diversity, the plasmids are not so variable that every isolate has a novel version of every plasmid (i.e., there appears to be a limited number of extant plasmid subtypes).
Conclusions: Although there have been many past recombination events, both homologous and nonhomologous, among the plasmids, particular organizational variants of these plasmids correlate with particular chromosomal genotypes, suggesting that there has not been rapid horizontal transfer of whole linear plasmids among B. burgdorferi lineages. We argue that plasmid rearrangements are essentially non-revertable and are present at a frequency of only about 0.65% that of single nucleotide changes, making rearrangement-derived novel junctions (mosaic boundaries) ideal phylogenetic markers in the study of B. burgdorferi population structure and plasmid evolution and exchange.
Keywords: B. burgdorferi; Genome rearrangement; Linear plasmid; Plasmid.
Figures
Similar articles
-
Primordial origin and diversification of plasmids in Lyme disease agent bacteria.BMC Genomics. 2018 Mar 27;19(1):218. doi: 10.1186/s12864-018-4597-x. BMC Genomics. 2018. PMID: 29580205 Free PMC article.
-
Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids.PLoS One. 2012;7(3):e33280. doi: 10.1371/journal.pone.0033280. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432010 Free PMC article.
-
Natural selection and recombination at host-interacting lipoprotein loci drive genome diversification of Lyme disease and related bacteria.mBio. 2024 Sep 11;15(9):e0174924. doi: 10.1128/mbio.01749-24. Epub 2024 Aug 15. mBio. 2024. PMID: 39145656 Free PMC article.
-
The plasmids of Borrelia burgdorferi: essential genetic elements of a pathogen.Plasmid. 2005 Jan;53(1):1-13. doi: 10.1016/j.plasmid.2004.10.006. Epub 2004 Dec 16. Plasmid. 2005. PMID: 15631949 Review.
-
vls Antigenic Variation Systems of Lyme Disease Borrelia: Eluding Host Immunity through both Random, Segmental Gene Conversion and Framework Heterogeneity.Microbiol Spectr. 2014 Dec;2(6):10.1128/microbiolspec.MDNA3-0038-2014. doi: 10.1128/microbiolspec.MDNA3-0038-2014. Microbiol Spectr. 2014. PMID: 26104445 Free PMC article. Review.
Cited by
-
High conservation combined with high plasticity: genomics and evolution of Borrelia bavariensis.BMC Genomics. 2020 Oct 8;21(1):702. doi: 10.1186/s12864-020-07054-3. BMC Genomics. 2020. PMID: 33032522 Free PMC article.
-
Genomic hybrid capture assay to detect Borrelia burgdorferi: an application to diagnose neuroborreliosis in horses.J Vet Diagn Invest. 2022 Sep;34(5):909-912. doi: 10.1177/10406387221112617. Epub 2022 Jul 21. J Vet Diagn Invest. 2022. PMID: 35864735 Free PMC article.
-
A Novel Rapid Sample Preparation Method for MALDI-TOF MS Permits Borrelia burgdorferi Sensu Lato Species and Isolate Differentiation.Front Microbiol. 2020 Apr 21;11:690. doi: 10.3389/fmicb.2020.00690. eCollection 2020. Front Microbiol. 2020. PMID: 32373099 Free PMC article.
-
Strict Conservation yet Non-Essential Nature of Plasmid Gene bba40 in the Lyme Disease Spirochete Borrelia burgdorferi.Microbiol Spectr. 2023 Jun 15;11(3):e0047723. doi: 10.1128/spectrum.00477-23. Epub 2023 Apr 3. Microbiol Spectr. 2023. PMID: 37010416 Free PMC article.
-
Bacteria have numerous distinctive groups of phage-plasmids with conserved phage and variable plasmid gene repertoires.Nucleic Acids Res. 2021 Mar 18;49(5):2655-2673. doi: 10.1093/nar/gkab064. Nucleic Acids Res. 2021. PMID: 33590101 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

