Diffusible substances from lactic acid bacterial cultures exert strong inhibitory effects on Listeria monocytogenes and Salmonella enterica serovar enteritidis in a co-culture model
- PMID: 28202007
- PMCID: PMC5312424
- DOI: 10.1186/s12866-017-0944-3
Diffusible substances from lactic acid bacterial cultures exert strong inhibitory effects on Listeria monocytogenes and Salmonella enterica serovar enteritidis in a co-culture model
Abstract
Background: Food-borne infections cause huge economic and human life losses. Listeria monocytogenes and Salmonella enterica serovar Enteritidis are among the top ranking pathogens causing such losses. Control of such infections is hampered by persistent contamination of foods and food-processing environments, resistance of pathogens to sanitizing agents, existence of heterogeneous populations of pathogens (including culturable and viable but non-culturable cells) within the same food items, and inability to detect all such pathogens by culture-based methods. Modern methods such as flow cytometry allow analyses of cells at the single cell level within a short time and enable better and faster detection of such pathogens and distinctions between live and dead cells. Such methods should be complemented by control strategies including the use of beneficial bacteria that produce metabolites capable of inhibiting food-borne pathogens. In this study, broth cultures of lactic acid bacteria (LAB) isolated from fermented milk were tested for production of substances capable of inhibiting L. monocytogenes and S. Enteritidis in co-culture with LAB by assessment of colony-forming units (CFU) and live:dead cell populations by flow cytometry.
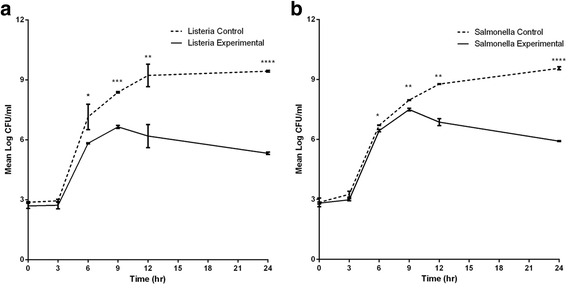
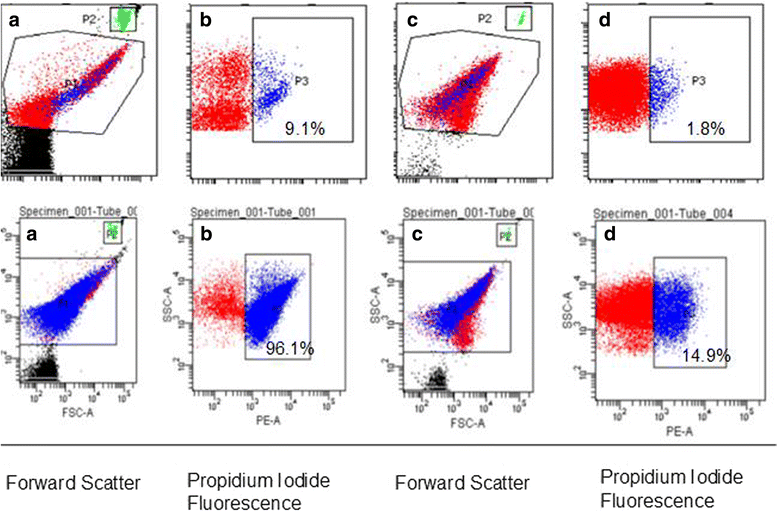
Results: The LAB isolates belonged to the species Lactococcus lactis, Enterococcus faecalis and Enterococcus faecium. Some LAB were effective in inhibition. Plating indicated up to 99% reduction in CFU from co-cultures compared to control cultures. Most of the bacteria in both cultures were in the viable but non-culturable state. The flow data showed that there were significantly higher dead cell numbers in co-cultures than in control cultures, indicating that such killing was caused by diffusible substances produced by the LAB cultures.
Conclusion: This study showed that metabolites from selected local LAB species can be used to significantly reduce pathogen load. However, conditions of use and application need to be further investigated and optimized for large-scale utilization.
Keywords: Co-culture; Inhibition; L. monocytogenes; Lactic acid bacteria; S. Enteritidis; Viable but nonculturable.
Figures



References
-
- Hitchins AD, Jinneman K. Detection and enumeration of Listeria monocytogenes in foods. Bacteriological analytical manual online. Washington: FDA; 2011.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

