Development of intron targeting (IT) markers specific for chromosome arm 4VS of Haynaldia villosa by chromosome sorting and next-generation sequencing
- PMID: 28202009
- PMCID: PMC5310052
- DOI: 10.1186/s12864-017-3567-z
Development of intron targeting (IT) markers specific for chromosome arm 4VS of Haynaldia villosa by chromosome sorting and next-generation sequencing
Abstract
Background: Haynaldia villosa (L.) Schur (syn. Dasypyrum villosum L. Candargy, 2n = 14, genome VV) is the tertiary gene pool of wheat, and thus a potential resource of genes for wheat improvement. Among other, wheat yellow mosaic (WYM) resistance gene Wss1 and a take-all resistance gene were identified on the short arm of chromosome 4 V (4VS) of H. villosa. We had obtained introgressions on 4VS chromosome arm, with the objective of utilizing the target genes. However, monitoring these introgressions has been a daunting task because of inadequate knowledge as to H.villosa genome, as reflected by the lack of specific markers.
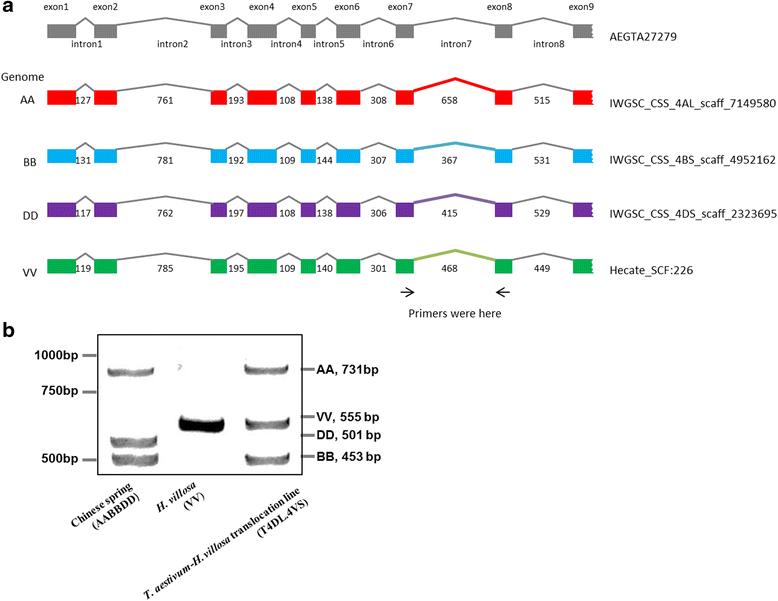
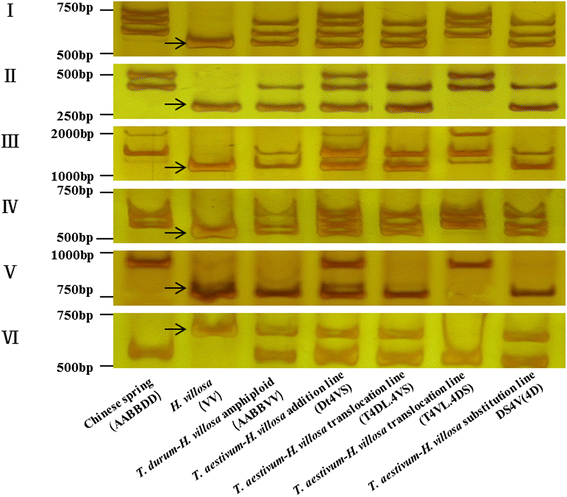
Results: This study aims to develop 4VS-specific markers by combination of chromosome sorting and next-generation sequencing. The short arm of chromosome 4VS of H.villosa was flow-sorted using a FACSVantage SE flow cytometer and sorter, and then sequenced by Illumina sequencing. The sequence of H. villosa 4VS was assembled by the software Hecate, and then was compared with the sequence assemblies of wheat chromosome arms 4AL, 4BS and 4DS and Ae. tauschii 4DS, with the objectives of identifying exon-exon junctions and localizing introns on chromosome 4VS of H. villosa. The intron length polymorphisms suitable for designing H. villosa primers were evaluated with criteria. Consequently, we designed a total of 359 intron targeting (IT) markers, among which 232 (64.62%) markers were specific for tracing the 4VS chromatin in the wheat background.
Conclusion: The combination of chromosome sorting and next-generation sequencing to develop specific IT markers for 4VS of H. villosa has high success rate and specificity, thus being applicable for the development of chromosome-specific markers for alien chromatin in wheat breeding.
Keywords: Haynaldia villosa; Intron polymorphism; Molecular marker; Triticum aestivum.
Figures
Similar articles
-
Sequencing flow-sorted short arm of Haynaldia villosa chromosome 4V provides insights into its molecular structure and virtual gene order.BMC Genomics. 2017 Oct 16;18(1):791. doi: 10.1186/s12864-017-4211-7. BMC Genomics. 2017. PMID: 29037165 Free PMC article.
-
Dissection and cytological mapping of chromosome arm 4VS by the development of wheat-Haynaldia villosa structural aberration library.Theor Appl Genet. 2020 Jan;133(1):217-226. doi: 10.1007/s00122-019-03452-8. Epub 2019 Oct 5. Theor Appl Genet. 2020. PMID: 31587088
-
Induction of 4VS chromosome recombinants using the CS ph1b mutant and mapping of the wheat yellow mosaic virus resistance gene from Haynaldia villosa.Theor Appl Genet. 2013 Dec;126(12):2921-30. doi: 10.1007/s00122-013-2181-y. Epub 2013 Aug 29. Theor Appl Genet. 2013. PMID: 23989649
-
Development of PCR markers specific to Dasypyrum villosum genome based on transcriptome data and their application in breeding Triticum aestivum-D. villosum#4 alien chromosome lines.BMC Genomics. 2019 Apr 15;20(1):289. doi: 10.1186/s12864-019-5630-4. BMC Genomics. 2019. PMID: 30987602 Free PMC article.
-
[Development and application of a genome specific PCR marker for Haynaldia villosa].Yi Chuan Xue Bao. 2003 Apr;30(4):350-6. Yi Chuan Xue Bao. 2003. PMID: 12812061 Chinese.
Cited by
-
Long-range assembly of sequences helps to unravel the genome structure and small variation of the wheat-Haynaldia villosa translocated chromosome 6VS.6AL.Plant Biotechnol J. 2021 Aug;19(8):1567-1578. doi: 10.1111/pbi.13570. Epub 2021 Mar 1. Plant Biotechnol J. 2021. PMID: 33606347 Free PMC article.
-
Utilization of the Dasypyrum genus for genetic improvement of wheat.Mol Breed. 2024 Dec 14;44(12):82. doi: 10.1007/s11032-024-01512-6. eCollection 2024 Dec. Mol Breed. 2024. PMID: 39687346 Review.
-
Fine mapping of wheat powdery mildew resistance gene Pm6 using 2B/2G homoeologous recombinants induced by the ph1b mutant.Theor Appl Genet. 2020 Apr;133(4):1265-1275. doi: 10.1007/s00122-020-03546-8. Epub 2020 Jan 23. Theor Appl Genet. 2020. PMID: 31974668
-
Flow Cytometric Analysis and Sorting of Plant Chromosomes.Methods Mol Biol. 2023;2672:177-200. doi: 10.1007/978-1-0716-3226-0_10. Methods Mol Biol. 2023. PMID: 37335476
-
Genetic Diversity and Evolutionary Analyses Reveal the Powdery Mildew Resistance Gene Pm21 Undergoing Diversifying Selection.Front Genet. 2020 May 12;11:489. doi: 10.3389/fgene.2020.00489. eCollection 2020. Front Genet. 2020. PMID: 32477413 Free PMC article.
References
-
- Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica. 1996;91(1):59–87.3. doi: 10.1007/BF00035277. - DOI
-
- Triebe B, Mukai Y, Dhaliwal HS, Martin TJ, Gill BS. Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and insitu hybridization. Theor Appl Genet. 1991;81(3):381–89. - PubMed
-
- Lukaszewski AJ, Lapinski B, Rybka K. Limitations of in situ hybridization with total genomic DNA in routine screening for alien introgressions in wheat. Cytogenet Genome Res. 2005;109(1–3):373–77. - PubMed
-
- Kimura M. Rare variant alleles in the light of the neutral theory. Mol Biol Evol. 1983;1(1):84–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

