HOXC8 regulates self-renewal, differentiation and transformation of breast cancer stem cells
- PMID: 28202042
- PMCID: PMC5312582
- DOI: 10.1186/s12943-017-0605-z
HOXC8 regulates self-renewal, differentiation and transformation of breast cancer stem cells
Abstract
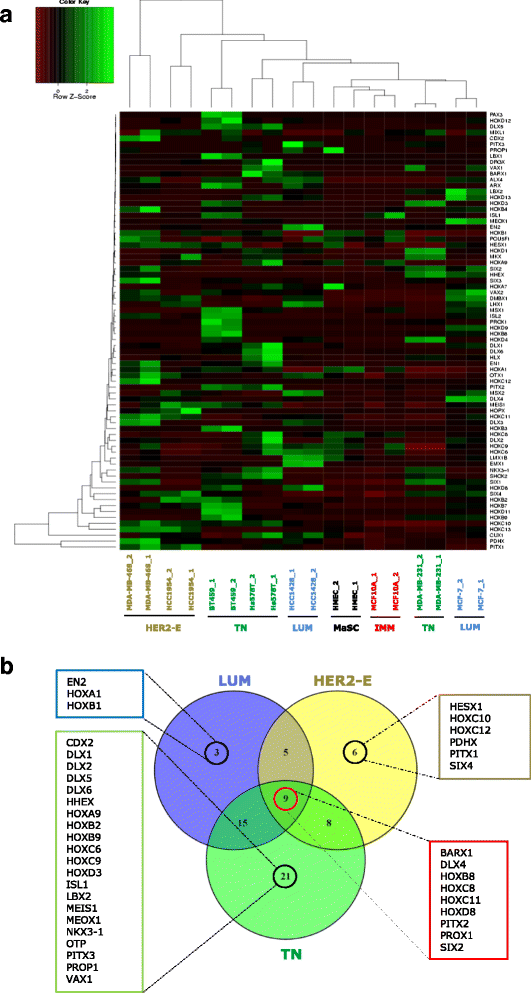
Background: Homeobox genes are master regulators of cell fate during embryonic development and their expression is altered in cancer. By regulating the balance between cell proliferation and differentiation, they maintain homeostasis of normal tissues. Here, we screened the expression of homeobox genes in mammary stem cells to establish their role in stem cells transformation in breast cancer.
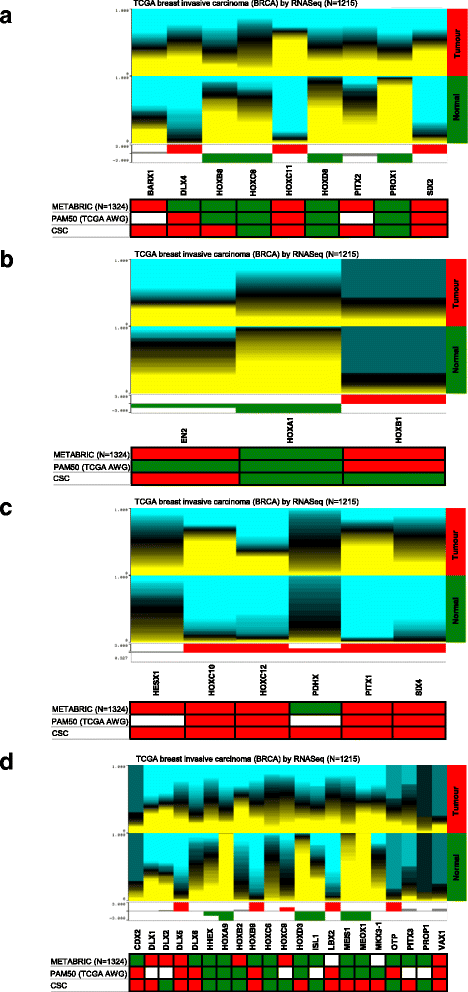
Methods: Using a Homeobox Genes PCR array, we screened 83 homeobox genes in normal cancer breast stem/progenitor cells isolated by flow cytometry. The candidate gene HOXC8 epigenetic regulation was studied by DNA methylation and miRNA expression analyses. Self-renewal and differentiation of HOXC8-overexpressing or knockdown cells were assessed by flow cytometry and mammosphere, 3D matrigel and soft agar assays. Clinical relevance of in vitro findings were validated by bioinformatics analysis of patient datasets from TCGA and METABRIC studies.
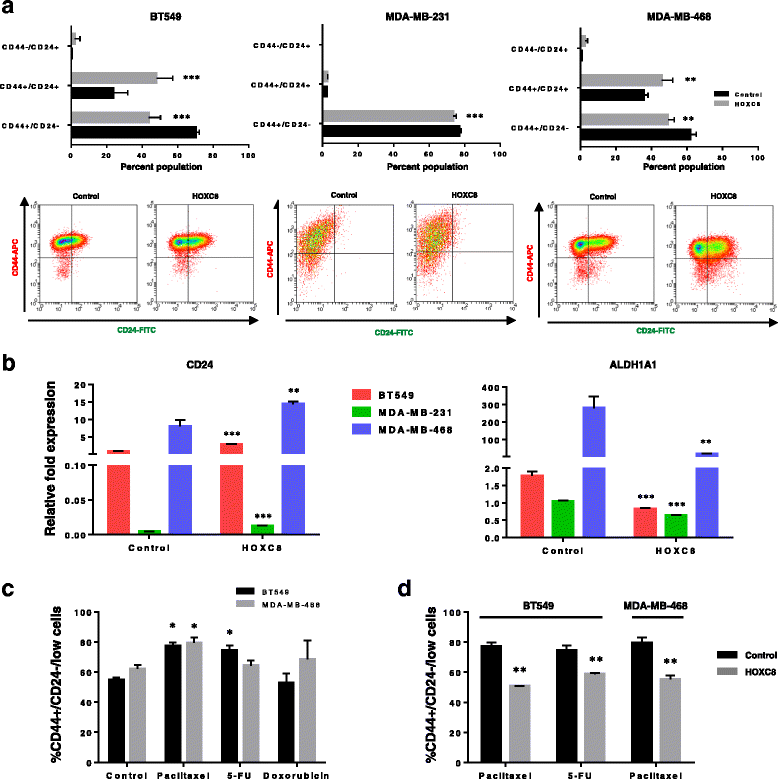
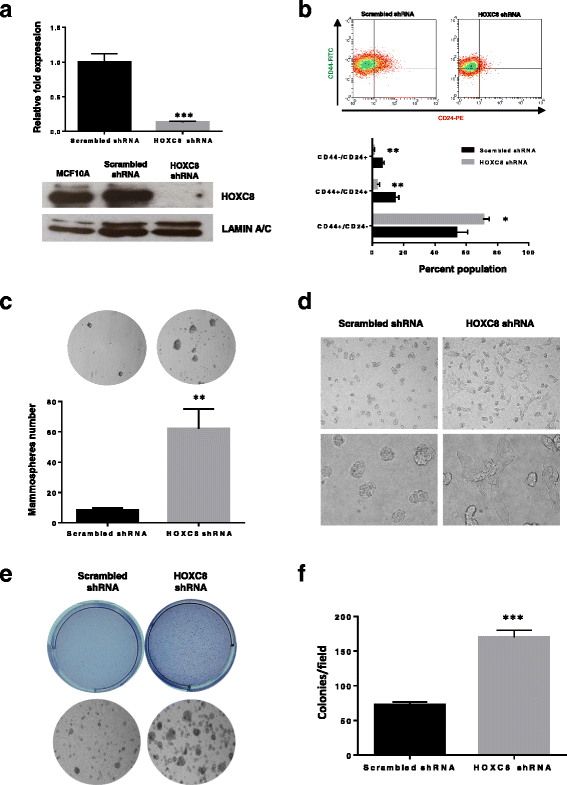
Results: In this study we demonstrate altered expression of homeobox genes in breast cancer stem/progenitor cells. HOXC8 was consistently downregulated in stem/progenitor cells of all breast molecular subtypes, thus representing an interesting tumour suppressor candidate. We show that downregulated expression of HOXC8 is associated with DNA methylation at the gene promoter and expression of miR196 family members. Functional studies demonstrated that HOXC8 gain of function induces a decrease in the CD44+/CD24-/low cancer stem cell population and proportion of chemoresistant cells, with a concomitant increase in CD24+ differentiated cells. Increased HOXC8 levels also decrease the ability of cancer cells to form mammospheres and to grow in anchorage-independent conditions. Furthermore, loss of HOXC8 in non-tumorigenic mammary epithelial cells expands the cancer stem/progenitor cells pool, increases stem cell self-renewal, prevents differentiation induced by retinoic acid and induces a transformed phenotype.
Conclusions: Taken together, our study points to an important role of homeobox genes in breast cancer stem/progenitor cell function and establishes HOXC8 as a suppressor of stemness and transformation in the mammary gland lineage.
Keywords: Breast cancer; Cancer stem cells; Differentiation; HOXC8; Homeobox genes; Self-renewal.
Figures




Similar articles
-
Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium.BMC Cancer. 2015 Sep 23;15:645. doi: 10.1186/s12885-015-1655-5. BMC Cancer. 2015. PMID: 26400441 Free PMC article.
-
Embigin, regulated by HOXC8, plays a suppressive role in breast tumorigenesis.Oncotarget. 2015 Sep 15;6(27):23496-509. doi: 10.18632/oncotarget.4360. Oncotarget. 2015. PMID: 26090721 Free PMC article.
-
[Tricostantin A inhibits self-renewal of breast cancer stem cells in vitro].Nan Fang Yi Ke Da Xue Xue Bao. 2013 Oct;33(10):1421-6. Nan Fang Yi Ke Da Xue Xue Bao. 2013. PMID: 24144739 Chinese.
-
Epigenetic and metabolic regulation of breast cancer stem cells.J Zhejiang Univ Sci B. 2015 Jan;16(1):10-7. doi: 10.1631/jzus.B1400172. J Zhejiang Univ Sci B. 2015. PMID: 25559951 Free PMC article. Review.
-
LncRNAs and microRNAs as Essential Regulators of Stemness in Breast Cancer Stem Cells.Biomolecules. 2021 Mar 3;11(3):380. doi: 10.3390/biom11030380. Biomolecules. 2021. PMID: 33802575 Free PMC article. Review.
Cited by
-
HOXD9 promotes the growth, invasion and metastasis of gastric cancer cells by transcriptional activation of RUFY3.J Exp Clin Cancer Res. 2019 Sep 23;38(1):412. doi: 10.1186/s13046-019-1399-1. J Exp Clin Cancer Res. 2019. PMID: 31547840 Free PMC article.
-
Retinoic Acids in the Treatment of Most Lethal Solid Cancers.J Clin Med. 2020 Jan 28;9(2):360. doi: 10.3390/jcm9020360. J Clin Med. 2020. PMID: 32012980 Free PMC article. Review.
-
Suppression of RNA editing by miR-17 inhibits the stemness of melanoma stem cells.Mol Ther Nucleic Acids. 2021 Dec 17;27:439-455. doi: 10.1016/j.omtn.2021.12.021. eCollection 2022 Mar 8. Mol Ther Nucleic Acids. 2021. PMID: 35036056 Free PMC article.
-
MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1.Stem Cell Res Ther. 2020 May 6;11(1):170. doi: 10.1186/s13287-020-01679-7. Stem Cell Res Ther. 2020. PMID: 32375892 Free PMC article.
-
A Case of Identity: HOX Genes in Normal and Cancer Stem Cells.Cancers (Basel). 2019 Apr 10;11(4):512. doi: 10.3390/cancers11040512. Cancers (Basel). 2019. PMID: 30974862 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

