Costs and benefits of natural transformation in Acinetobacter baylyi
- PMID: 28202049
- PMCID: PMC5312590
- DOI: 10.1186/s12866-017-0953-2
Costs and benefits of natural transformation in Acinetobacter baylyi
Abstract
Background: Natural transformation enables acquisition of adaptive traits and drives genome evolution in prokaryotes. Yet, the selective forces responsible for the evolution and maintenance of natural transformation remain elusive since taken-up DNA has also been hypothesized to provide benefits such as nutrients or templates for DNA repair to individual cells.
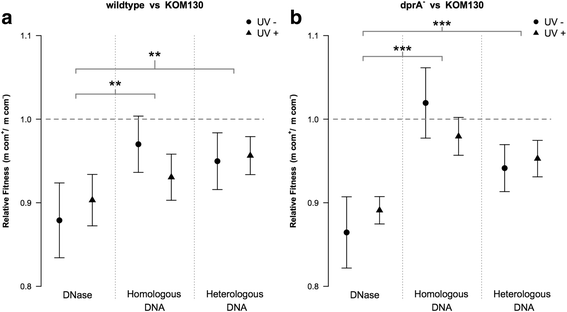
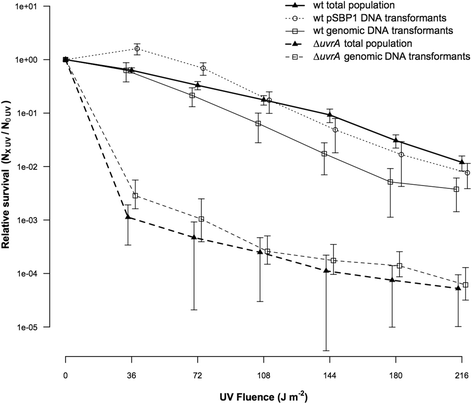
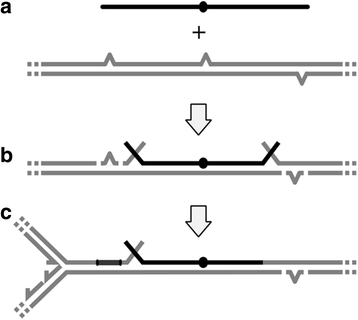
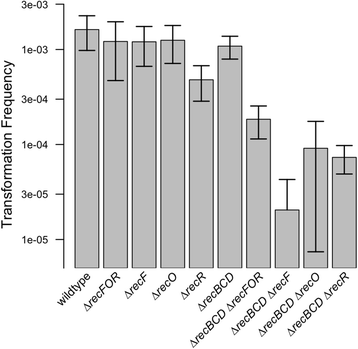
Results: We investigated the immediate effects of DNA uptake and recombination on the naturally competent bacterium Acinetobacter baylyi in both benign and genotoxic conditions. In head-to-head competition experiments between DNA uptake-proficient and -deficient strains, we observed a fitness benefit of DNA uptake independent of UV stress. This benefit was found with both homologous and heterologous DNA and was independent of recombination. Recombination with taken-up DNA reduced survival of transformed cells with increasing levels of UV-stress through interference with nucleotide excision repair, suggesting that DNA strand breaks occur during recombination attempts with taken-up DNA. Consistent with this, we show that absence of RecBCD and RecFOR recombinational DNA repair pathways strongly decrease natural transformation.
Conclusions: Our data show a physiological benefit of DNA uptake unrelated to recombination. In contrast, recombination during transformation is a strand break inducing process that represents a previously unrecognized cost of natural transformation.
Keywords: Bacterial evolution; Competence; DNA repair; DprA; Horizontal gene transfer; Natural transformation.
Figures
Similar articles
-
Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi.Microbiology (Reading). 2007 Jul;153(Pt 7):2259-2270. doi: 10.1099/mic.0.2007/005256-0. Microbiology (Reading). 2007. PMID: 17600070
-
The recombination initiation functions DprA and RecFOR suppress microindel mutations in Acinetobacter baylyi ADP1.Mol Microbiol. 2024 Jul;122(1):1-10. doi: 10.1111/mmi.15277. Epub 2024 May 17. Mol Microbiol. 2024. PMID: 38760330
-
The RecBCD and SbcCD DNases suppress homology-facilitated illegitimate recombination during natural transformation of Acinetobacter baylyi.Microbiology (Reading). 2008 Aug;154(Pt 8):2437-2445. doi: 10.1099/mic.0.2008/018382-0. Microbiology (Reading). 2008. PMID: 18667576
-
Single-strand gap repair involves both RecF and RecBCD pathways.Curr Genet. 2016 Aug;62(3):519-21. doi: 10.1007/s00294-016-0575-5. Epub 2016 Feb 13. Curr Genet. 2016. PMID: 26874520 Review.
-
Acinetobacter baylyi ADP1: transforming the choice of model organism.IUBMB Life. 2011 Dec;63(12):1075-80. doi: 10.1002/iub.530. Epub 2011 Oct 27. IUBMB Life. 2011. PMID: 22034222 Review.
Cited by
-
Testing for the fitness benefits of natural transformation during community-embedded evolution.Microbiology (Reading). 2023 Aug;169(8):001375. doi: 10.1099/mic.0.001375. Microbiology (Reading). 2023. PMID: 37526972 Free PMC article.
-
Effect of Nutritional Determinants and TonB on the Natural Transformation of Riemerella anatipestifer.Front Microbiol. 2021 Aug 10;12:644868. doi: 10.3389/fmicb.2021.644868. eCollection 2021. Front Microbiol. 2021. PMID: 34447355 Free PMC article.
-
Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria.Nat Rev Microbiol. 2017 Oct;15(10):621-629. doi: 10.1038/nrmicro.2017.66. Epub 2017 Jul 10. Nat Rev Microbiol. 2017. PMID: 28690319 Review.
-
Molecular mechanisms and applications of natural transformation in bacteria.Front Microbiol. 2025 Jun 24;16:1578813. doi: 10.3389/fmicb.2025.1578813. eCollection 2025. Front Microbiol. 2025. PMID: 40630188 Free PMC article. Review.
-
Distribution of fitness effects of cross-species transformation reveals potential for fast adaptive evolution.ISME J. 2023 Jan;17(1):130-139. doi: 10.1038/s41396-022-01325-5. Epub 2022 Oct 12. ISME J. 2023. PMID: 36224268 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

