The Crumbs_C isoform of Drosophila shows tissue- and stage-specific expression and prevents light-dependent retinal degeneration
- PMID: 28202468
- PMCID: PMC5312091
- DOI: 10.1242/bio.020040
The Crumbs_C isoform of Drosophila shows tissue- and stage-specific expression and prevents light-dependent retinal degeneration
Abstract
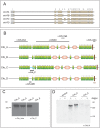
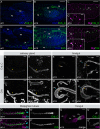
Drosophila Crumbs (Crb) is a key regulator of epithelial polarity and fulfils a plethora of other functions, such as growth regulation, morphogenesis of photoreceptor cells and prevention of retinal degeneration. This raises the question how a single gene regulates such diverse functions, which in mammals are controlled by three different paralogs. Here, we show that in Drosophila different Crb protein isoforms are differentially expressed as a result of alternative splicing. All isoforms are transmembrane proteins that differ by just one EGF-like repeat in their extracellular portion. Unlike Crb_A, which is expressed in most embryonic epithelia from early stages onward, Crb_C is expressed later and only in a subset of embryonic epithelia. Flies specifically lacking Crb_C are homozygous viable and fertile. Strikingly, these flies undergo light-dependent photoreceptor degeneration despite the fact that the other isoforms are expressed and properly localised at the stalk membrane. This allele now provides an ideal possibility to further unravel the molecular mechanisms by which Drosophila crb protects photoreceptor cells from the detrimental consequences of light-induced cell stress.
Keywords: Alternative splicing; EGF-like repeat; Epithelial polarity; Mutually exclusive exon.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures





Similar articles
-
PAR-Complex and Crumbs Function During Photoreceptor Morphogenesis and Retinal Degeneration.Front Cell Neurosci. 2018 Mar 29;12:90. doi: 10.3389/fncel.2018.00090. eCollection 2018. Front Cell Neurosci. 2018. PMID: 29651238 Free PMC article. Review.
-
Crumbs regulates polarity and prevents light-induced degeneration of the simple eyes of Drosophila, the ocelli.Eur J Cell Biol. 2012 Sep;91(9):706-16. doi: 10.1016/j.ejcb.2012.03.006. Epub 2012 May 18. Eur J Cell Biol. 2012. PMID: 22608020
-
Drosophila Lin-7 is a component of the Crumbs complex in epithelia and photoreceptor cells and prevents light-induced retinal degeneration.Eur J Cell Biol. 2008 Mar;87(3):123-36. doi: 10.1016/j.ejcb.2007.11.002. Epub 2008 Jan 4. Eur J Cell Biol. 2008. PMID: 18177979
-
Unique cell biological profiles of retinal disease-causing missense mutations in the polarity protein Crumbs.J Cell Sci. 2017 Jul 1;130(13):2147-2158. doi: 10.1242/jcs.197178. Epub 2017 May 17. J Cell Sci. 2017. PMID: 28515229
-
[Role of Crumbs proteins in the control of epithelial cell and photoreceptor morphogenesis].Med Sci (Paris). 2004 Jun-Jul;20(6-7):663-7. doi: 10.1051/medsci/2004206-7663. Med Sci (Paris). 2004. PMID: 15329816 Review. French.
Cited by
-
The apical protein Apnoia interacts with Crumbs to regulate tracheal growth and inflation.PLoS Genet. 2019 Jan 15;15(1):e1007852. doi: 10.1371/journal.pgen.1007852. eCollection 2019 Jan. PLoS Genet. 2019. PMID: 30645584 Free PMC article.
-
Loss of Crb2b-lf leads to anterior segment defects in old zebrafish.Biol Open. 2020 Feb 11;9(2):bio047555. doi: 10.1242/bio.047555. Biol Open. 2020. PMID: 31988089 Free PMC article.
-
PAR-Complex and Crumbs Function During Photoreceptor Morphogenesis and Retinal Degeneration.Front Cell Neurosci. 2018 Mar 29;12:90. doi: 10.3389/fncel.2018.00090. eCollection 2018. Front Cell Neurosci. 2018. PMID: 29651238 Free PMC article. Review.
-
Crumbs organizes the transport machinery by regulating apical levels of PI(4,5)P2 in Drosophila.Elife. 2019 Nov 7;8:e50900. doi: 10.7554/eLife.50900. Elife. 2019. PMID: 31697234 Free PMC article.
-
Changes in endolysosomal organization define a pre-degenerative state in the crumbs mutant Drosophila retina.PLoS One. 2019 Dec 13;14(12):e0220220. doi: 10.1371/journal.pone.0220220. eCollection 2019. PLoS One. 2019. PMID: 31834921 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

