A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence
- PMID: 28202493
- PMCID: PMC5376759
- DOI: 10.15252/emmm.201607336
A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence
Abstract
Klebsiella pneumoniae is an important cause of multidrug-resistant infections worldwide. Recent studies highlight the emergence of multidrug-resistant K. pneumoniae strains which show resistance to colistin, a last-line antibiotic, arising from mutational inactivation of the mgrB regulatory gene. However, the precise molecular resistance mechanisms of mgrB-associated colistin resistance and its impact on virulence remain unclear. Here, we constructed an mgrB gene K. pneumoniae mutant and performed characterisation of its lipid A structure, polymyxin and antimicrobial peptide resistance, virulence and inflammatory responses upon infection. Our data reveal that mgrB mutation induces PhoPQ-governed lipid A remodelling which confers not only resistance to polymyxins, but also enhances K. pneumoniae virulence by decreasing antimicrobial peptide susceptibility and attenuating early host defence response activation. Overall, our findings have important implications for patient management and antimicrobial stewardship, while also stressing antibiotic resistance development is not inexorably linked with subdued bacterial fitness and virulence.
Keywords: Klebsiella pneumoniae; mgrB; antimicrobial peptides; polymyxins; virulence.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

Growth kinetics of K. pneumoniae 51245 (blue and grey) and 52145‐ΔmgrB strains (red and black) cultured in LB broth (LB) and 2% glucose M9 minimal media supplemented with thiamine and MgSO4 (M9) over 24 h at 37°C. Values are presented as the mean ± SD of three independent experiments measured in triplicate.
Short‐term biofilm assay results for the K. pneumoniae 52145, 52145‐ΔmgrB and 52145‐ΔmgrBCom strains. Results are displayed as per cent biofilm relative to the wild‐type strain with values presented as the mean ± SD of three independent experiments. Level of significance versus 52145 (**P = 0.007) was determined using two‐way unpaired t‐test.
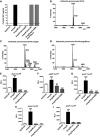
- A
Minimal inhibitory concentrations to colistin of the K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom, 52145‐ΔmgrB‐ΔphoQGB, 52145‐ΔmgrB‐ΔpmrAB and 52145‐ΔmgrB‐ΔphoQGB‐phoPQCom strains. The broken line represents the European Committee on Antimicrobial Susceptibility Testing MIC breakpoint.
- B–D
Negative ion MALDI‐TOF mass spectrometry spectra of lipid A purified from (B) K. pneumoniae 52145, (C) 52145‐ΔmgrB and (D) 52145‐ΔmgrBCom strains. Data represent the mass‐to‐charge (m/z) ratios of each lipid A species detected and are representative of three extractions.
- E–I
Activity of the lpxO (E), pagP (F), pmrC (G) and pmrH (H) and phoP (I) promoters in K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom, 52145‐ΔmgrB‐ΔphoQGB carrying lucFF transcriptional fusions. Values (expressed in relative luminescence units) are presented as the mean ± SD of three independent experiments measured in triplicate. ***P < 0.0005; **P = 0.0071; *P = 0.019; versus 52145 determined using two‐way unpaired t‐test.

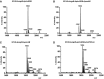
- A–D
Negative ion MALDI‐TOF mass spectrometry spectra of lipid A isolated from the K. pneumoniae 52145‐ΔmgrBΔphoQGB (A), 52145‐ΔmgrB‐ΔphoQGB‐ΔpmrAB (B), 52145‐ΔmgrBΔpmrAB (C) and 52145‐ΔmgrB‐ΔphoQGB‐phoPQCom (D) strains. Data represent the mass‐to‐charge (m/z) ratios of each lipid A species detected and are representative of three extractions.
- A–D
Per cent survival of K. pneumoniae 52145, 52145‐ΔmgrB and 52145‐ΔmgrBCom following 1‐h exposure to the following: (A) human neutrophil peptide‐1 (1.2 μM), (B) human β‐defensin‐1 (3 μM), (C) human β‐defensin‐2 (3 μM) and (D) human β‐defensin‐3 (7 μM). Values are presented as the mean ± SD of three independent experiments measured in duplicate. ***P = 0.0006; **P = 0.01–0.001; *P = 0.013; versus Kp52145 determined using two‐way unpaired t‐test.
- E
Resistance of K. pneumoniae 52145, 52145‐ΔmgrB and 52145‐ΔmgrBCom to antimicrobial factors produced by G. mellonella after 24‐h exposure to 106 heat‐killed E. coli cells. The experiments were undertaken using a radial diffusion bioassay with data expressed as radial diffusion units (10 units = 1 mm). Values are presented as the mean ± SD of three independent experiments measured in triplicate. ***P < 0.0001; versus 52145 determined using two‐way unpaired t‐test.
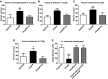
- A, B
Etest® minimal inhibitory concentrations to colistin and polymyxin B of K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom compared to the double, triple and quadruple 52145‐ΔmgrB mutant strains.
- C, D
Per cent survival of the K. pneumoniae 52145 wild‐type and mutant strains after exposure to 20 μg/ml colistin (C) and polymyxin B (D) over 1 h. Values are presented as the mean ± SD of three independent experiments measured in duplicate. ***P < 0.0001; *P = 0.0195; versus 52145‐ΔmgrB determined using a two‐way unpaired t‐test.
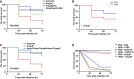
- A–C
Kaplan–Meier plots showing the per cent survival of G. mellonella over 72 h post‐infection with 105 organisms of the following: (A) K. pneumoniae 52145 (blue), 52145‐ΔmgrB (red), 52145‐ΔmgrBCom (green) and 52145‐ΔmgrB‐ΔphoQGB (grey), (B) clinical K. pneumoniae strains T1a (blue) and T1b (red), and (C) K. pneumoniae 52145 (blue), 52145‐ΔmgrB (red) and 52145‐ΔpmrC‐ΔlpxO‐ΔmgrB‐ΔpmrF‐ΔpagP quintuple‐mutant (green) strains. Forty larvae were infected in each group. Level of significance was determined using the log‐rank (Mantel–Cox) test with Bonferroni correction for multiple comparisons where applicable [α = (A) 0.0008; (B) 0.05; (C) 0.017]. P‐values presented correspond to the difference between (A) 52145‐ΔmgrB and the other 52145 strains and (B) T1a versus T1b.
- D
Survival curve of G. mellonella primed with 106 heat‐killed E. coli cells (HKEc) or phosphate‐buffered saline (PBS) and then exposed to PBS (black, grey) or 106 organisms of K. pneumoniae 52145 (blue, magenta) and 52145‐ΔmgrB (red, green) over 72 h. Ten larvae were infected in each group. Level of significance was determined using the log‐rank (Mantel–Cox) test with Bonferroni correction for multiple comparisons (α = 0.008).

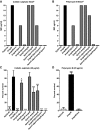
Kaplan–Meier plot showing the per cent survival of G. mellonella over 72 h post‐infection with 106 organisms of K. pneumoniae 52145‐ΔmanC (blue) and 52145‐ΔmgrB‐ΔmanC (red). Forty larvae were infected in each group. Level of significance was determined using the log‐rank (Mantel–Cox) test (α = 0.05).
Activity of the cps promoter in K. pneumoniae 52145 and 52145‐ΔmgrB carrying the cps::lucFF transcriptional fusion. Values (expressed in relative luminescence units [540 nm]) are presented as the mean ± SD of three independent experiments measured in triplicate. Level of significance was determined using two‐way unpaired t‐test.
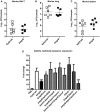
- A–C
Bacterial loads of K. pneumoniae 52145 and 52145‐ΔmgrB in nasal‐associated lymphoid tissue (NALT), lung and spleen homogenates of infected mice after 24 h. Six mice per group were infected with log10 colony‐forming unit values presented as the mean ± SD. Data were analysed using two‐way unpaired t‐test.
- D
Expression of lysozyme produced by G. mellonella after 8 h of infection with K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom, 52145‐ΔmgrB‐ΔphoQGB, 52145‐ΔpmrC‐ΔlpxO‐ΔmgrB, 52145‐ΔmgrB‐ΔlpxO‐ΔpmrF, 52145‐ΔpmrC‐ΔlpxO‐ΔmgrB‐ΔpmrF, 52145‐ΔlpxO, 52145‐ΔmgrB‐ΔlpxO and 52145‐ΔmgrB‐ΔlpxO‐lpxOCom as determined by reverse transcriptase quantitative real‐time PCR. Three larvae per group were infected, and values are presented as the mean ± SD of two independent cDNA preparations measured in duplicate. ***P < 0.0001; *P = 0.035; versus 52145 determined using two‐way unpaired t‐test.
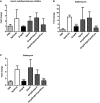
- A–C
G. mellonella antimicrobial peptide expression determined after 8 h of infection with K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom, 52145‐ΔlpxO, 52145‐ΔmgrB‐ΔlpxO and 52145‐ΔmgrB‐ΔlpxO‐lpxOCom by reverse transcriptase quantitative real‐time PCR. Three larvae per group were infected, and values are presented as the mean ± SD of two independent cDNA preparations measured in duplicate. ***P < 0.0001; versus 52145 determined using two‐way unpaired t‐test.
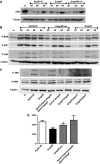
Immunoblot analysis of IκBα and tubulin levels in lysates of iBMDM cells infected with K. pneumoniae 52145, 52145‐ΔmgrB and 52145‐ΔmgrBCom for the indicated times.
Immunoblot analysis of phospho‐ERK (P‐ERK), phospho‐p38 (P‐p38), phospho‐JNK (P‐JNK) and tubulin levels in lysates of iBMDMs cells infected with K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom and for the indicated times.
Immunoblot analysis of phospho‐ERK (P‐ERK), phospho‐JNK (P‐JNK) and tubulin levels in lysates of iBMDMs cells infected with K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔpmrC‐ΔlpxO‐ΔmgrB‐ΔpmrF‐ΔpagP, 52145‐ΔmgrB‐ΔpmrC, 52145‐ΔmgrB‐ΔpagP, 52145‐ΔmgrB‐ΔpmrF and 52145‐ΔmgrB‐ΔlpxO for 40 min.
TNF‐α secretion by iBMDM macrophages stimulated for 6 h with 1 × 105 UV‐killed K. pneumoniae 52145, 52145‐ΔmgrB, 52145‐ΔmgrBCom and 52145‐ΔpmrC‐ΔlpxO‐ΔmgrB‐ΔpmrF‐ΔpagP. ***P < 0.0001; *P = 0.02; versus 52145 determined using two‐way unpaired t‐test (mean ± SD).
Similar articles
-
MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae.Cells. 2022 Sep 26;11(19):2995. doi: 10.3390/cells11192995. Cells. 2022. PMID: 36230959 Free PMC article. Review.
-
Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors.Infect Immun. 2011 Sep;79(9):3718-32. doi: 10.1128/IAI.05226-11. Epub 2011 Jun 27. Infect Immun. 2011. PMID: 21708987 Free PMC article.
-
The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae.J Antimicrob Chemother. 2015 Jan;70(1):75-80. doi: 10.1093/jac/dku323. Epub 2014 Sep 3. J Antimicrob Chemother. 2015. PMID: 25190723
-
MgrB-Dependent Colistin Resistance in Klebsiella pneumoniae Is Associated with an Increase in Host-to-Host Transmission.mBio. 2022 Apr 26;13(2):e0359521. doi: 10.1128/mbio.03595-21. Epub 2022 Mar 21. mBio. 2022. PMID: 35311534 Free PMC article.
-
The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae.Int J Environ Res Public Health. 2020 Aug 28;17(17):6278. doi: 10.3390/ijerph17176278. Int J Environ Res Public Health. 2020. PMID: 32872324 Free PMC article. Review.
Cited by
-
MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae.Cells. 2022 Sep 26;11(19):2995. doi: 10.3390/cells11192995. Cells. 2022. PMID: 36230959 Free PMC article. Review.
-
Insights into Kinases of ESKAPE Pathogens for Therapeutic Interventions.Cardiovasc Hematol Agents Med Chem. 2024;22(3):276-297. doi: 10.2174/0118715257267497231128093529. Cardiovasc Hematol Agents Med Chem. 2024. PMID: 39403051 Review.
-
Antibacterial Activity of LCB10-0200 against Klebsiella pneumoniae.Antibiotics (Basel). 2021 Sep 29;10(10):1185. doi: 10.3390/antibiotics10101185. Antibiotics (Basel). 2021. PMID: 34680766 Free PMC article.
-
Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae.Microb Genom. 2018 Mar;4(3):e000158. doi: 10.1099/mgen.0.000158. Epub 2018 Feb 12. Microb Genom. 2018. PMID: 29431605 Free PMC article.
-
Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains.Microb Genom. 2020 Dec;6(12):mgen000474. doi: 10.1099/mgen.0.000474. Epub 2020 Nov 10. Microb Genom. 2020. PMID: 33170117 Free PMC article.
References
-
- Afacan NJ, Janot LM, Hancock REW (2013) Host defense peptides: immune modulation and antimicrobial activity in vivo In Antimicrobial peptides and innate immunity, Hiemstra PS, Zaat SAJ. (eds), pp 321–358. New York: Springer;
-
- Aubert D, Hamad M, Valvano M (2014) A markerless deletion method for genetic manipulation of Burkholderia cenocepacia and other multidrug‐resistant gram‐negative bacteria In Host‐bacteria interactions, Vergunst AC, O'Callaghan D. (eds), pp 311–327. New York: Springer; - PubMed
-
- Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM et al (2011) Phosphoethanolamine modification of lipid A in colistin‐resistant variants of Acinetobacter baumannii mediated by the pmrAB two‐component regulatory system. Antimicrob Agents Chemother 55: 3370–3379 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

