Nucleosome-like, Single-stranded DNA (ssDNA)-Histone Octamer Complexes and the Implication for DNA Double Strand Break Repair
- PMID: 28202543
- PMCID: PMC5392674
- DOI: 10.1074/jbc.M117.776369
Nucleosome-like, Single-stranded DNA (ssDNA)-Histone Octamer Complexes and the Implication for DNA Double Strand Break Repair
Abstract
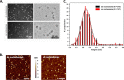
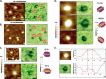
Repair of DNA double strand breaks (DSBs) is key for maintenance of genome integrity. When DSBs are repaired by homologous recombination, DNA ends can undergo extensive processing, producing long stretches of single-stranded DNA (ssDNA). In vivo, DSB processing occurs in the context of chromatin, and studies indicate that histones may remain associated with processed DSBs. Here we demonstrate that histones are not evicted from ssDNA after in vitro chromatin resection. In addition, we reconstitute histone-ssDNA complexes (termed ssNucs) with ssDNA and recombinant histones and analyze these particles by a combination of native gel electrophoresis, sedimentation velocity, electron microscopy, and a recently developed electrostatic force microscopy technique, DREEM (
Keywords: DNA repair; DSB processing; analytical ultracentrifugation; atomic force microscopy (AFM); chromatin remodeling; nucleosome; ssDNA.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Khanna K. K., and Jackson S. P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

