Exploring targeted therapy of osteosarcoma using proteomics data
- PMID: 28203090
- PMCID: PMC5295800
- DOI: 10.2147/OTT.S119993
Exploring targeted therapy of osteosarcoma using proteomics data
Abstract
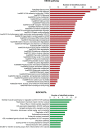
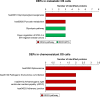
Despite multimodal therapeutic treatments of osteosarcoma (OS), some patients develop resistance to currently available regimens and eventually end up with recurrent or metastatic outcomes. Many attempts have been made to discover effective drugs for improving outcome; however, due to the heterogeneity of the disease, new therapeutic options have not yet been identified. This study aims to explore potential targeted therapy related to protein profiles of OS. In this review of proteomics studies, we extracted data on differentially expressed proteins (DEPs) from archived literature in PubMed and our in-house repository. The data were divided into three experimental groups, DEPs in 1) OS/OB: OS vs osteoblastic (OB) cells, 2) metastasis: metastatic vs non-metastatic sublines plus fresh tissues from primary OS with and without pulmonary metastasis, and 3) chemoresistance: spheroid (higher chemoresistance) vs monolayer cells plus fresh tissues from biopsies from good and poor responders. All up-regulated protein entities in the list of DEPs were sorted and cross-referenced with identifiers of targets of US Food and Drug Administration (FDA)-approved agents and chemical inhibitors. We found that many targets of FDA-approved antineoplastic agents, mainly a group of epigenetic regulators, kinases, and proteasomes, were highly expressed in OS cells. Additionally, some overexpressed proteins were targets of FDA-approved non-cancer drugs, including immunosuppressive and antiarrhythmic drugs. The resulting list of chemical agents showed that some transferase enzyme inhibitors might have anticancer activity. We also explored common targets of OS/OB and metastasis groups, including amidophosphoribosyltransferase (PPAT), l-lactate dehydrogenase B chain (LDHB), and pyruvate kinase M2 (PKM2) as well as the common target of all categories, cathepsin D (CTSD). This study demonstrates the benefits of a text mining approach to exploring therapeutic targets related to protein expression patterns. These results suggest possible repurposing of some FDA-approved medicines for the treatment of OS and using chemical inhibitors in drug screening tests.
Keywords: FDA-approved drugs; osteosarcoma; proteomics; targeted therapy; text mining.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures



Similar articles
-
Candidate cancer-targeting agents identified by expression-profiling arrays.Onco Targets Ther. 2013 Apr 23;6:447-58. doi: 10.2147/OTT.S42858. Print 2013. Onco Targets Ther. 2013. PMID: 23637543 Free PMC article.
-
New drug candidates for osteosarcoma: Drug repurposing based on gene expression signature.Comput Biol Med. 2021 Jul;134:104470. doi: 10.1016/j.compbiomed.2021.104470. Epub 2021 May 7. Comput Biol Med. 2021. PMID: 34004576
-
Properties of FDA-approved small molecule protein kinase inhibitors.Pharmacol Res. 2019 Jun;144:19-50. doi: 10.1016/j.phrs.2019.03.006. Epub 2019 Mar 13. Pharmacol Res. 2019. PMID: 30877063 Review.
-
Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy.Eur J Pharm Biopharm. 2015 Jun;93:52-79. doi: 10.1016/j.ejpb.2015.03.018. Epub 2015 Mar 23. Eur J Pharm Biopharm. 2015. PMID: 25813885 Review.
-
Tegavivint and the β-Catenin/ALDH Axis in Chemotherapy-Resistant and Metastatic Osteosarcoma.J Natl Cancer Inst. 2019 Nov 1;111(11):1216-1227. doi: 10.1093/jnci/djz026. J Natl Cancer Inst. 2019. PMID: 30793158 Free PMC article.
Cited by
-
Molecular Biology of Osteosarcoma.Cancers (Basel). 2020 Jul 31;12(8):2130. doi: 10.3390/cancers12082130. Cancers (Basel). 2020. PMID: 32751922 Free PMC article. Review.
-
Circ-ATAD1 is overexpressed in osteosarcoma (OS) and suppresses the maturation of miR-154-5p to increase cell invasion and migration.J Orthop Surg Res. 2021 Dec 2;16(1):699. doi: 10.1186/s13018-021-02809-4. J Orthop Surg Res. 2021. PMID: 34857012 Free PMC article.
-
Phosphoribosyl Pyrophosphate Amidotransferase Promotes the Progression of Thyroid Cancer via Regulating Pyruvate Kinase M2.Onco Targets Ther. 2020 Aug 3;13:7629-7639. doi: 10.2147/OTT.S253137. eCollection 2020. Onco Targets Ther. 2020. PMID: 32801776 Free PMC article.
-
Mass Spectrometric-Based Proteomics for Biomarker Discovery in Osteosarcoma: Current Status and Future Direction.Int J Mol Sci. 2022 Aug 28;23(17):9741. doi: 10.3390/ijms23179741. Int J Mol Sci. 2022. PMID: 36077137 Free PMC article. Review.
-
Integration of genomic copy number variations and chemotherapy-response biomarkers in pediatric sarcoma.BMC Med Genomics. 2019 Jan 31;12(Suppl 1):23. doi: 10.1186/s12920-018-0456-5. BMC Med Genomics. 2019. PMID: 30704460 Free PMC article.
References
-
- Settakorn J, Lekawanvijit S, Arpornchayanon O, et al. Spectrum of bone tumors in Chiang Mai University Hospital, Thailand according to WHO classification 2002: a study of 1,001 cases. J Med Assoc Thai. 2006;89(6):780–787. - PubMed
-
- Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: a meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ) 2015;44(12):547–553. - PubMed
-
- Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

