Ribosomal RNA Genes Contribute to the Formation of Pseudogenes and Junk DNA in the Human Genome
- PMID: 28204512
- PMCID: PMC5381670
- DOI: 10.1093/gbe/evw307
Ribosomal RNA Genes Contribute to the Formation of Pseudogenes and Junk DNA in the Human Genome
Abstract
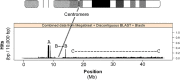
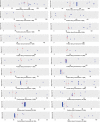
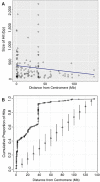
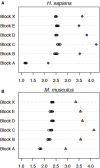
Approximately 35% of the human genome can be identified as sequence devoid of a selected-effect function, and not derived from transposable elements or repeated sequences. We provide evidence supporting a known origin for a fraction of this sequence. We show that: 1) highly degraded, but near full length, ribosomal DNA (rDNA) units, including both 45S and Intergenic Spacer (IGS), can be found at multiple sites in the human genome on chromosomes without rDNA arrays, 2) that these rDNA sequences have a propensity for being centromere proximal, and 3) that sequence at all human functional rDNA array ends is divergent from canonical rDNA to the point that it is pseudogenic. We also show that small sequence strings of rDNA (from 45S + IGS) can be found distributed throughout the genome and are identifiable as an "rDNA-like signal", representing 0.26% of the q-arm of HSA21 and ∼2% of the total sequence of other regions tested. The size of sequence strings found in the rDNA-like signal intergrade into the size of sequence strings that make up the full-length degrading rDNA units found scattered throughout the genome. We conclude that the displaced and degrading rDNA sequences are likely of a similar origin but represent different stages in their evolution towards random sequence. Collectively, our data suggests that over vast evolutionary time, rDNA arrays contribute to the production of junk DNA. The concept that the production of rDNA pseudogenes is a by-product of concerted evolution represents a previously under-appreciated process; we demonstrate here its importance.
Keywords: concerted evolution; degraded rDNA; genome evolution; vestigial centromere.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





Similar articles
-
rRNA Pseudogenes in Filamentous Ascomycetes as Revealed by Genome Data.G3 (Bethesda). 2017 Aug 7;7(8):2695-2703. doi: 10.1534/g3.117.044016. G3 (Bethesda). 2017. PMID: 28637809 Free PMC article.
-
Polymorphism and evolution of ribosomal DNA in tea (Camellia sinensis, Theaceae).Mol Phylogenet Evol. 2015 Aug;89:63-72. doi: 10.1016/j.ympev.2015.03.020. Epub 2015 Apr 11. Mol Phylogenet Evol. 2015. PMID: 25871774
-
A Portrait of Ribosomal DNA Contacts with Hi-C Reveals 5S and 45S rDNA Anchoring Points in the Folded Human Genome.Genome Biol Evol. 2016 Dec 31;8(11):3545-3558. doi: 10.1093/gbe/evw257. Genome Biol Evol. 2016. PMID: 27797956 Free PMC article.
-
Integrative rDNAomics-Importance of the Oldest Repetitive Fraction of the Eukaryote Genome.Genes (Basel). 2019 May 7;10(5):345. doi: 10.3390/genes10050345. Genes (Basel). 2019. PMID: 31067804 Free PMC article. Review.
-
Annotating non-coding regions of the genome.Nat Rev Genet. 2010 Aug;11(8):559-71. doi: 10.1038/nrg2814. Epub 2010 Jul 13. Nat Rev Genet. 2010. PMID: 20628352 Review.
Cited by
-
Transcriptional Mutagenesis Prevents Ribosomal DNA Deterioration: The Role of Duplications and Deletions.Genome Biol Evol. 2019 Nov 1;11(11):3207-3217. doi: 10.1093/gbe/evz235. Genome Biol Evol. 2019. PMID: 31651950 Free PMC article.
-
Role of Pseudogenes in Tumorigenesis.Cancers (Basel). 2018 Aug 1;10(8):256. doi: 10.3390/cancers10080256. Cancers (Basel). 2018. PMID: 30071685 Free PMC article. Review.
-
Effective ribosomal RNA depletion for single-cell total RNA-seq by scDASH.PeerJ. 2021 Jan 15;9:e10717. doi: 10.7717/peerj.10717. eCollection 2021. PeerJ. 2021. PMID: 33520469 Free PMC article.
-
Methylation of 45S Ribosomal DNA (rDNA) Is Associated with Cancer and Aging in Humans.Int J Genomics. 2021 Jan 28;2021:8818007. doi: 10.1155/2021/8818007. eCollection 2021. Int J Genomics. 2021. PMID: 33575316 Free PMC article. Review.
-
Structure and biochemistry-guided engineering of an all-RNA system for DNA insertion with R2 retrotransposons.Nat Commun. 2025 Jul 2;16(1):6079. doi: 10.1038/s41467-025-61321-z. Nat Commun. 2025. PMID: 40603868 Free PMC article.
References
-
- Adler D. 1992. Idiogram album: mouse. http://pathology.washington.edu/research/cytopages/.
-
- Adler D, Willis M. 1991. Idiogram album: human. http://pathology.washington.edu/research/cytopages/.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
-
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 89:247–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

