Epigenetic signatures of invasive status in populations of marine invertebrates
- PMID: 28205577
- PMCID: PMC5311950
- DOI: 10.1038/srep42193
Epigenetic signatures of invasive status in populations of marine invertebrates
Abstract
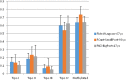
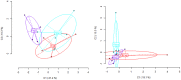
Epigenetics, as a DNA signature that affects gene expression and enables rapid reaction of an organism to environmental changes, is likely involved in the process of biological invasions. DNA methylation is an epigenetic mechanism common to plants and animals for regulating gene expression. In this study we show, for the first time in any marine species, significant reduction of global methylation levels during the expansive phase of a pygmy mussel (Xenostrobus securis) recent invasion in Europe (two-year old), while in older introductions such epigenetic signature of invasion was progressively reduced. Decreased methylation was interpreted as a rapid way of increasing phenotypic plasticity that would help invasive populations to thrive. This epigenetic signature of early invasion was stronger than the expected environmental signature of environmental stress in younger populations sampled from ports, otherwise detected in a much older population (>90 year old) of the also invasive tubeworm Ficopomatus enigmaticus established in similar locations. Higher epigenetic than genetic diversity found in X. securis was confirmed from F. enigmaticus samples. As reported for introduced plants and vertebrates, epigenetic variation could compensate for relatively lower genetic variation caused by founder effects. These phenomena were compared with epigenetic mechanisms involved in metastasis, as parallel processes of community (biological invasion) and organism (cancer) invasions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Bossdorf O., Richards C. L. & Pigliucci M. Epigenetics for ecologists. Ecol. Lett. 11, 106–115 (2008). - PubMed
-
- Kardong K. V. Epigenomics: the new science of functional and evolutionary morphology. Anim. Biol. 53, 225–243 (2003).
-
- Zhang Y. Y., Fischer M., Colot V. & Bossdorf O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 197, 314–22 (2013). - PubMed
-
- Suarez-Ulloa V., Gonzalez-Romero R. & Eirin-Lopez J. M. Environmental epigenetics: A promising venue for developing next-generation pollution biomonitoring tools in marine invertebrates. Mar. Poll. Bull. 98, 5–13 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

