Aspartate Glycosylation Triggers Isomerization to Isoaspartate
- PMID: 28207246
- PMCID: PMC5431074
- DOI: 10.1021/jacs.6b12866
Aspartate Glycosylation Triggers Isomerization to Isoaspartate
Abstract
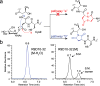
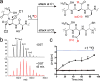
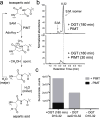
O-Linked β-N-acetylglucosamine transferase (OGT) is an essential human enzyme that glycosylates numerous nuclear and cytoplasmic proteins on serine and threonine. It also cleaves Host cell factor 1 (HCF-1) by a mechanism in which the first step involves glycosylation on glutamate. Replacing glutamate with aspartate in an HCF-1 proteolytic repeat was shown to prevent peptide backbone cleavage, but whether aspartate glycosylation occurred was not examined. We report here that OGT glycosylates aspartate much faster than it glycosylates glutamate in an otherwise identical model peptide substrate; moreover, once formed, the glycosyl aspartate reacts further to form a succinimide intermediate that hydrolyzes to produce the corresponding isoaspartyl peptide. Aspartate-to-isoaspartate isomerization in proteins occurs in cells but was previously thought to be exclusively non-enzymatic. Our findings suggest it may also be enzyme-catalyzed. In addition to OGT, enzymes that may catalyze aspartate to isoaspartate isomerization include PARPs, enzymes known to ribosylate aspartate residues in the process of poly(ADP-ribosyl)ation.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
The conserved threonine-rich region of the HCF-1PRO repeat activates promiscuous OGT:UDP-GlcNAc glycosylation and proteolysis activities.J Biol Chem. 2018 Nov 16;293(46):17754-17768. doi: 10.1074/jbc.RA118.004185. Epub 2018 Sep 17. J Biol Chem. 2018. PMID: 30224358 Free PMC article.
-
How the glycosyltransferase OGT catalyzes amide bond cleavage.Nat Chem Biol. 2016 Nov;12(11):899-901. doi: 10.1038/nchembio.2173. Epub 2016 Sep 12. Nat Chem Biol. 2016. PMID: 27618188 Free PMC article.
-
HCF-1 is cleaved in the active site of O-GlcNAc transferase.Science. 2013 Dec 6;342(6163):1235-9. doi: 10.1126/science.1243990. Science. 2013. PMID: 24311690 Free PMC article.
-
The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells?Annu Rev Biochem. 2016 Jun 2;85:631-57. doi: 10.1146/annurev-biochem-060713-035344. Annu Rev Biochem. 2016. PMID: 27294441 Review.
-
The making of a sweet modification: structure and function of O-GlcNAc transferase.J Biol Chem. 2014 Dec 12;289(50):34424-32. doi: 10.1074/jbc.R114.604405. Epub 2014 Oct 21. J Biol Chem. 2014. PMID: 25336649 Free PMC article. Review.
Cited by
-
O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix.J Am Chem Soc. 2018 Mar 14;140(10):3510-3513. doi: 10.1021/jacs.7b13546. Epub 2018 Mar 5. J Am Chem Soc. 2018. PMID: 29485866 Free PMC article.
-
Deciphering the Properties and Functions of Glycoproteins Using Quantitative Proteomics.J Proteome Res. 2023 Jun 2;22(6):1571-1588. doi: 10.1021/acs.jproteome.3c00015. Epub 2023 Apr 3. J Proteome Res. 2023. PMID: 37010087 Free PMC article. Review.
-
O-Methyltransferase-Mediated Incorporation of a β-Amino Acid in Lanthipeptides.J Am Chem Soc. 2019 Oct 23;141(42):16790-16801. doi: 10.1021/jacs.9b07396. Epub 2019 Oct 15. J Am Chem Soc. 2019. PMID: 31568727 Free PMC article.
-
Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges.Biochemistry. 2018 Jan 16;57(2):177-185. doi: 10.1021/acs.biochem.7b00861. Epub 2017 Nov 3. Biochemistry. 2018. PMID: 29064683 Free PMC article. Review.
-
An Unusually Rapid Protein Backbone Modification Stabilizes the Essential Bacterial Enzyme MurA.Biochemistry. 2020 Oct 6;59(39):3683-3695. doi: 10.1021/acs.biochem.0c00502. Epub 2020 Sep 24. Biochemistry. 2020. PMID: 32930597 Free PMC article.
References
-
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W. Cell. 2011;144:376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

