Structure, kinetic characterization and subcellular localization of the two ribulose 5-phosphate epimerase isoenzymes from Trypanosoma cruzi
- PMID: 28207833
- PMCID: PMC5312968
- DOI: 10.1371/journal.pone.0172405
Structure, kinetic characterization and subcellular localization of the two ribulose 5-phosphate epimerase isoenzymes from Trypanosoma cruzi
Abstract
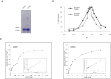
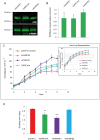
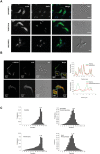
The enzyme of the pentose phosphate pathway (PPP) ribulose-5-phosphate-epimerase (RPE) is encoded by two genes present in the genome of Trypanosoma cruzi CL Brener clone: TcRPE1 and TcRPE2. Despite high sequence similarity at the amino acid residue level, the recombinant isoenzymes show a strikingly different kinetics. Whereas TcRPE2 follows a typical michaelian behavior, TcRPE1 shows a complex kinetic pattern, displaying a biphasic curve, suggesting the coexistence of -at least- two kinetically different molecular forms. Regarding the subcellular localization in epimastigotes, whereas TcRPE1 is a cytosolic enzyme, TcRPE2 is localized in glycosomes. To our knowledge, TcRPE2 is the first PPP isoenzyme that is exclusively localized in glycosomes. Over-expression of TcRPE1, but not of TcRPE2, significantly reduces the parasite doubling time in vitro, as compared with wild type epimastigotes. Both TcRPEs represent single domain proteins exhibiting the classical α/β TIM-barrel fold, as expected for enzymes with this activity. With regard to the architecture of the active site, all the important amino acid residues for catalysis -with the exception of M58- are also present in both TcRPEs models. The superimposition of the binding pocket of both isoenzyme models shows that they adopt essentially identical positions in the active site with a residue specific RMSD < 2Å, with the sole exception of S12, which displays a large deviation (residue specific RMSD: 11.07 Å). Studies on the quaternary arrangement of these isoenzymes reveal that both are present in a mixture of various oligomeric species made up of an even number of molecules, probably pointing to the dimer as their minimal functional unit. This multiplicity of oligomeric species has not been reported for any of the other RPEs studied so far and it might bear implications for the regulation of TcRPEs activity, although further investigation will be necessary to unravel the physiological significance of these structural findings.
Conflict of interest statement
Figures



References
-
- Barrett MP. The Pentose Phosphate Pathway and Parasitic Protozoa. Parasitol Today. 1997;13(I):11–6. - PubMed
-
- Wood T. Purification and properties of d-ribulose-5-phosphate 3-epimerase from calf liver. BBA—Enzymol. 1979;570(2):352–62. - PubMed
-
- Nowitzki U, Wyrich R, Westhoff P, Henze K, Schnarrenberger C, Martin W. Cloning of the amphibolic Calvin cycle/OPPP enzyme d-ribulose-5-phosphate 3-epimerase (EC 5.1.3.1) from spinach chloroplasts: functional and evolutionary aspects. Plant Mol Biol. 1995;29(6):1279–91. - PubMed
-
- Bär J, Naumann M, Reuter R, Kopperschläger G. Improved purification of ribulose 5-phosphate 3-epimerase from Saccharomyces cerevisiae and characterization of the enzyme. Bioseparation. 1996;6(4):233–41. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

