Novel approaches for the taxonomic and metabolic characterization of lactobacilli: Integration of 16S rRNA gene sequencing with MALDI-TOF MS and 1H-NMR
- PMID: 28207855
- PMCID: PMC5312945
- DOI: 10.1371/journal.pone.0172483
Novel approaches for the taxonomic and metabolic characterization of lactobacilli: Integration of 16S rRNA gene sequencing with MALDI-TOF MS and 1H-NMR
Abstract
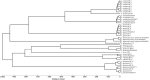
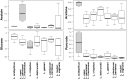
Lactobacilli represent a wide range of bacterial species with several implications for the human host. They play a crucial role in maintaining the ecological equilibrium of different biological niches and are essential for fermented food production and probiotic formulation. Despite the consensus about the 'health-promoting' significance of Lactobacillus genus, its genotypic and phenotypic characterization still poses several difficulties. The aim of this study was to assess the integration of different approaches, genotypic (16S rRNA gene sequencing), proteomic (MALDI-TOF MS) and metabolomic (1H-NMR), for the taxonomic and metabolic characterization of Lactobacillus species. For this purpose we analyzed 40 strains of various origin (intestinal, vaginal, food, probiotics), belonging to different species. The high discriminatory power of MALDI-TOF for species identification was underlined by the excellent agreement with the genotypic analysis. Indeed, MALDI-TOF allowed to correctly identify 39 out of 40 Lactobacillus strains at the species level, with an overall concordance of 97.5%. In the perspective to simplify the MALDI TOF sample preparation, especially for routine practice, we demonstrated the perfect agreement of the colony-picking from agar plates with the protein extraction protocol. 1H-NMR analysis, applied to both culture supernatants and bacterial lysates, identified a panel of metabolites whose variations in concentration were associated with the taxonomy, but also revealed a high intra-species variability that did not allow a species-level identification. Therefore, despite not suitable for mere taxonomic purposes, metabolomics can be useful to correlate particular biological activities with taxonomy and to understand the mechanisms related to the antimicrobial effect shown by some Lactobacillus species.
Conflict of interest statement
Figures


Similar articles
-
Rapid species-level identification of vaginal and oral lactobacilli using MALDI-TOF MS analysis and 16S rDNA sequencing.BMC Microbiol. 2014 Dec 14;14:312. doi: 10.1186/s12866-014-0312-5. BMC Microbiol. 2014. PMID: 25495549 Free PMC article.
-
16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry.BMC Microbiol. 2016 Jun 13;16:105. doi: 10.1186/s12866-016-0732-5. BMC Microbiol. 2016. PMID: 27296852 Free PMC article.
-
Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools.J Dairy Sci. 2018 Feb;101(2):979-991. doi: 10.3168/jds.2017-13642. Epub 2017 Nov 15. J Dairy Sci. 2018. PMID: 29153517
-
Taxonomy of Lactobacilli and Bifidobacteria.Curr Issues Intest Microbiol. 2007 Sep;8(2):44-61. Curr Issues Intest Microbiol. 2007. PMID: 17542335 Review.
-
The Genomic Basis of Lactobacilli as Health-Promoting Organisms.Microbiol Spectr. 2017 Jun;5(3):10.1128/microbiolspec.bad-0011-2016. doi: 10.1128/microbiolspec.BAD-0011-2016. Microbiol Spectr. 2017. PMID: 28643623 Free PMC article. Review.
Cited by
-
Diet, Microbiota, and Gut Permeability-The Unknown Triad in Rheumatoid Arthritis.Front Med (Lausanne). 2018 Dec 14;5:349. doi: 10.3389/fmed.2018.00349. eCollection 2018. Front Med (Lausanne). 2018. PMID: 30619860 Free PMC article. Review.
-
Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens.Poult Sci. 2018 Mar 1;97(3):930-936. doi: 10.3382/ps/pex396. Poult Sci. 2018. PMID: 29294082 Free PMC article.
-
A Snapshot, Using a Multi-Omic Approach, of the Metabolic Cross-Talk and the Dynamics of the Resident Microbiota in Ripening Cheese Inoculated with Listeria innocua.Foods. 2024 Jun 18;13(12):1912. doi: 10.3390/foods13121912. Foods. 2024. PMID: 38928853 Free PMC article.
-
Diversity of vaginal microbiome and metabolome during genital infections.Sci Rep. 2019 Oct 1;9(1):14095. doi: 10.1038/s41598-019-50410-x. Sci Rep. 2019. PMID: 31575935 Free PMC article.
-
Exopolysaccharides from vaginal lactobacilli modulate microbial biofilms.Microb Cell Fact. 2023 Mar 8;22(1):45. doi: 10.1186/s12934-023-02053-x. Microb Cell Fact. 2023. PMID: 36890519 Free PMC article.
References
-
- Felis GE, Dellaglio F. Taxonomy of lactobacilli and bifidobacteria. Curr Issues Intest Microbiol. 2007;8: 44–61. - PubMed
-
- Axelsson L. Lactic acid bacteria: classification and physiology In: Salminen S, von Wright A, Ouwehand A, editors. Lactic acid bacteria: microbiological and functional aspects, 3rd ed Marcel Dekker, New York, NY; 2003. pp. 1–66.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

