Analysis of acquired mutations in transgenes arising in Ba/F3 transformation assays: findings and recommendations
- PMID: 28208123
- PMCID: PMC5355038
- DOI: 10.18632/oncotarget.15392
Analysis of acquired mutations in transgenes arising in Ba/F3 transformation assays: findings and recommendations
Abstract
The identification and functional validation of potentially oncogenic mutations in leukemia is an essential step toward a future of personalized targeted therapy. To assess the oncogenic capacity of individual mutations, reliable and scalable in vitro experimental approaches are required. Since 1988, researchers have used the IL-3 dependent Ba/F3 transformation assay to validate the oncogenic potential of mutations to drive factor-independent growth. Here we report a previously unrecognized phenomenon whereby Ba/F3 cells, engineered to express weakly transforming mutations, present with additional acquired mutations in the expressed transgene following factor withdrawal. Using four mutations with known transformative capacity in three cytokine receptors (CSF2RB, CSF3R and IL7R), we demonstrate that the mutated receptors are highly susceptible to acquiring additional mutations. These acquired mutations of unknown functional significance are selected by factor withdrawal but appear to exist prior to the removal of growth factor. This anomaly has the potential to confound efforts to both validate and characterize oncogenic mutations in leukemia, particularly when it is not standard practice to sequence validate cDNAs from transformed Ba/F3 lines. We present specific recommendations to detect and mitigate this phenomenon in future research using Ba/F3 transformation assays, along with methods to make the Ba/F3 assay more quantitative.
Keywords: Ba/F3 transformation assay; functional validation; leukemia; oncogenes; reproducibility in research.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
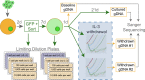
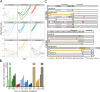


Comment in
-
Unwanted acquired mutations in Ba/F3 transformation assays.Oncotarget. 2017 Mar 28;8(13):20523-20524. doi: 10.18632/oncotarget.16268. Oncotarget. 2017. PMID: 28423556 Free PMC article. No abstract available.
References
-
- Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science (New York, NY) 1986;233(4760):212–214. - PubMed
-
- Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41(3):727–734. - PubMed
-
- Daley GQ, McLaughlin J, Witte ON, Baltimore D. The CML-specific P210 bcr/abl protein, unlike v-abl, does not transform NIH/3T3 fibroblasts. Science (New York, NY) 1987;237(4814):532–535. - PubMed
-
- Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Current opinion in oncology. 2007;19(1):55–60. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

