Elaborated Action of the Human Primosome
- PMID: 28208743
- PMCID: PMC5333051
- DOI: 10.3390/genes8020062
Elaborated Action of the Human Primosome
Abstract
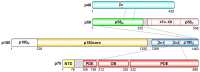
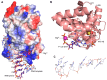
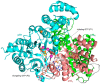
The human primosome is a 340-kilodalton complex of primase (DNA-dependent RNA polymerase) and DNA polymerase α, which initiates genome replication by synthesizing chimeric RNA-DNA primers for DNA polymerases δ and ϵ. Accumulated biochemical and structural data reveal the complex mechanism of concerted primer synthesis by two catalytic centers. First, primase generates an RNA primer through three steps: initiation, consisting of dinucleotide synthesis from two nucleotide triphosphates; elongation, resulting in dinucleotide extension; and termination, owing to primase inhibition by a mature 9-mer primer. Then Polα, which works equally well on DNA:RNA and DNA:DNA double helices, intramolecularly catches the template primed by a 9mer RNA and extends the primer with dNTPs. All primosome transactions are highly coordinated by autoregulation through the alternating activation/inhibition of the catalytic centers. This coordination is mediated by the small C-terminal domain of the primase accessory subunit, which forms a tight complex with the template:primer, shuttles between the primase and DNA polymerase active sites, and determines their access to the substrate.
Keywords: DNA polymerase α; DNA replication; RNA synthesis; human; initiation; primase; primosome; protein-DNA interaction; steric hindrance; termination.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



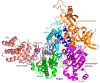


References
-
- Pellegrini L. The pol α-primase complex. Subcell. Biochem. 2012;62:157–169. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

