Dynamics of p53: A Master Decider of Cell Fate
- PMID: 28208785
- PMCID: PMC5333055
- DOI: 10.3390/genes8020066
Dynamics of p53: A Master Decider of Cell Fate
Abstract
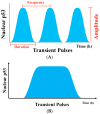
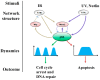
Cellular stress-induced temporal alterations-i.e., dynamics-are typically exemplified by the dynamics of p53 that serve as a master to determine cell fate. p53 dynamics were initially identified as the variations of p53 protein levels. However, a growing number of studies have shown that p53 dynamics are also manifested in variations in the activity, spatial location, and posttranslational modifications of p53 proteins, as well as the interplay among all p53 dynamical features. These are essential in determining a specific outcome of cell fate. In this review, we discuss the importance of the multifaceted features of p53 dynamics and their roles in the cell fate decision process, as well as their potential applications in p53-based cancer therapy. The review provides new insights into p53 signaling pathways and their potentials in the development of new strategies in p53-based cancer therapy.
Keywords: p53 dynamics; cell fate decision; cell signaling network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Impact of the adenoviral E4 Orf3 protein on the activity and posttranslational modification of p53.J Virol. 2015 Mar;89(6):3209-20. doi: 10.1128/JVI.03072-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568206 Free PMC article.
-
Dynamics of P53 in response to DNA damage: Mathematical modeling and perspective.Prog Biophys Mol Biol. 2015 Nov;119(2):175-82. doi: 10.1016/j.pbiomolbio.2015.08.017. Epub 2015 Aug 13. Prog Biophys Mol Biol. 2015. PMID: 26278643 Review.
-
Modeling the interplay between the HIF-1 and p53 pathways in hypoxia.Sci Rep. 2015 Sep 8;5:13834. doi: 10.1038/srep13834. Sci Rep. 2015. PMID: 26346319 Free PMC article.
-
Decision making by p53: life, death and cancer.Cell Death Differ. 2003 Apr;10(4):431-42. doi: 10.1038/sj.cdd.4401183. Cell Death Differ. 2003. PMID: 12719720 Review.
-
From structure to dynamics: frequency tuning in the p53-Mdm2 network I. Logical approach.J Theor Biol. 2009 Jun 21;258(4):561-77. doi: 10.1016/j.jtbi.2009.02.005. Epub 2009 Feb 21. J Theor Biol. 2009. PMID: 19233211
Cited by
-
Inhibition of mycobacteria proliferation in macrophages by low cisplatin concentration through phosphorylated p53-related apoptosis pathway.PLoS One. 2023 Jan 31;18(1):e0281170. doi: 10.1371/journal.pone.0281170. eCollection 2023. PLoS One. 2023. PMID: 36719870 Free PMC article.
-
Network Theory Analysis of Allosteric Drug-Rescue Mechanisms in the Tumor Suppressor Protein p53 Y220C Mutant.Int J Mol Sci. 2025 Jul 17;26(14):6884. doi: 10.3390/ijms26146884. Int J Mol Sci. 2025. PMID: 40725138 Free PMC article.
-
ROS: Basic Concepts, Sources, Cellular Signaling, and its Implications in Aging Pathways.Oxid Med Cell Longev. 2022 Oct 19;2022:1225578. doi: 10.1155/2022/1225578. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36312897 Free PMC article. Review.
-
The Activation of the Tumor Suppressor Protein p53 by Acriflavine Leads to Mitochondrial Dysfunction and Improves the Radiosensitivity of Colon Cancer Cells.J Immunol Res. 2022 Jul 29;2022:1328542. doi: 10.1155/2022/1328542. eCollection 2022. J Immunol Res. 2022. PMID: 35935580 Free PMC article.
-
Exploring the mechanism of aloe-emodin in the treatment of liver cancer through network pharmacology and cell experiments.Front Pharmacol. 2023 Oct 12;14:1238841. doi: 10.3389/fphar.2023.1238841. eCollection 2023. Front Pharmacol. 2023. PMID: 37900162 Free PMC article.
References
-
- Miyashita T., Reed J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

