Making life difficult for Clostridium difficile: augmenting the pathogen's metabolic model with transcriptomic and codon usage data for better therapeutic target characterization
- PMID: 28209199
- PMCID: PMC5314682
- DOI: 10.1186/s12918-017-0395-3
Making life difficult for Clostridium difficile: augmenting the pathogen's metabolic model with transcriptomic and codon usage data for better therapeutic target characterization
Abstract
Background: Clostridium difficile is a bacterium which can infect various animal species, including humans. Infection with this bacterium is a leading healthcare-associated illness. A better understanding of this organism and the relationship between its genotype and phenotype is essential to the search for an effective treatment. Genome-scale metabolic models contain all known biochemical reactions of a microorganism and can be used to investigate this relationship.
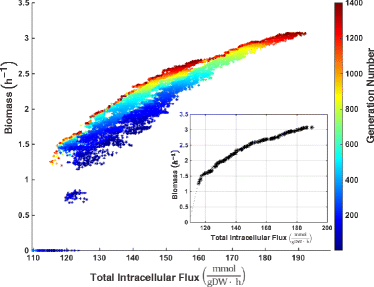
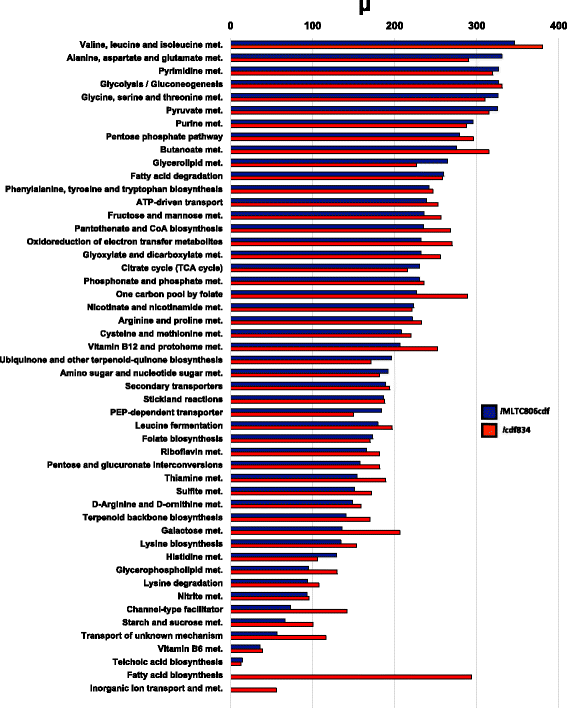
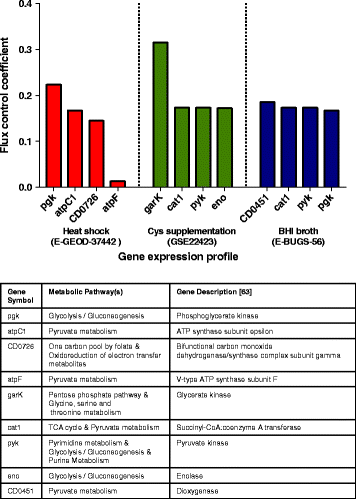
Results: We present icdf834, an updated metabolic network of C. difficile that builds on iMLTC806cdf and features 1227 reactions, 834 genes, and 807 metabolites. We used this metabolic network to reconstruct the metabolic landscape of this bacterium. The standard metabolic model cannot account for changes in the bacterial metabolism in response to different environmental conditions. To account for this limitation, we also integrated transcriptomic data, which details the gene expression of the bacterium in a wide array of environments. Importantly, to bridge the gap between gene expression levels and protein abundance, we accounted for the synonymous codon usage bias of the bacterium in the model. To our knowledge, this is the first time codon usage has been quantified and integrated into a metabolic model. The metabolic fluxes were defined as a function of protein abundance. To determine potential therapeutic targets using the model, we conducted gene essentiality and metabolic pathway sensitivity analyses and calculated flux control coefficients. We obtained 92.3% accuracy in predicting gene essentiality when compared to experimental data for C. difficile R20291 (ribotype 027) homologs. We validated our context-specific metabolic models using sensitivity and robustness analyses and compared model predictions with literature on C. difficile. The model predicts interesting facets of the bacterium's metabolism, such as changes in the bacterium's growth in response to different environmental conditions.
Conclusions: After an extensive validation process, we used icdf834 to obtain state-of-the-art predictions of therapeutic targets for C. difficile. We show how context-specific metabolic models augmented with codon usage information can be a beneficial resource for better understanding C. difficile and for identifying novel therapeutic targets. We remark that our approach can be applied to investigate and treat against other pathogens.
Keywords: Antibiotic resistance; Clostridium difficile; Flux balance analysis; Genome scale modeling; Metabolic modeling; Metabolic networks; Metabolic pathways; Sensitivity analysis.
Figures

Similar articles
-
A curated C. difficile strain 630 metabolic network: prediction of essential targets and inhibitors.BMC Syst Biol. 2014 Oct 15;8:117. doi: 10.1186/s12918-014-0117-z. BMC Syst Biol. 2014. PMID: 25315994 Free PMC article.
-
The (p)ppGpp Synthetase RSH Mediates Stationary-Phase Onset and Antibiotic Stress Survival in Clostridioides difficile.J Bacteriol. 2020 Sep 8;202(19):e00377-20. doi: 10.1128/JB.00377-20. Print 2020 Sep 8. J Bacteriol. 2020. PMID: 32661079 Free PMC article.
-
High-throughput analysis of gene essentiality and sporulation in Clostridium difficile.mBio. 2015 Feb 24;6(2):e02383. doi: 10.1128/mBio.02383-14. mBio. 2015. PMID: 25714712 Free PMC article.
-
Antimicrobial resistance in Clostridioides difficile.Eur J Clin Microbiol Infect Dis. 2021 Dec;40(12):2459-2478. doi: 10.1007/s10096-021-04311-5. Epub 2021 Aug 24. Eur J Clin Microbiol Infect Dis. 2021. PMID: 34427801 Review.
-
The current riboswitch landscape in Clostridioides difficile.Microbiology (Reading). 2024 Oct;170(10):001508. doi: 10.1099/mic.0.001508. Microbiology (Reading). 2024. PMID: 39405103 Free PMC article. Review.
Cited by
-
Predictive regulatory and metabolic network models for systems analysis of Clostridioides difficile.Cell Host Microbe. 2021 Nov 10;29(11):1709-1723.e5. doi: 10.1016/j.chom.2021.09.008. Epub 2021 Oct 11. Cell Host Microbe. 2021. PMID: 34637780 Free PMC article.
-
The Genome-Scale Integrated Networks in Microorganisms.Front Microbiol. 2018 Feb 23;9:296. doi: 10.3389/fmicb.2018.00296. eCollection 2018. Front Microbiol. 2018. PMID: 29527198 Free PMC article. Review.
-
A diurnal flux balance model of Synechocystis sp. PCC 6803 metabolism.PLoS Comput Biol. 2019 Jan 24;15(1):e1006692. doi: 10.1371/journal.pcbi.1006692. eCollection 2019 Jan. PLoS Comput Biol. 2019. PMID: 30677028 Free PMC article.
-
Human Systems Biology and Metabolic Modelling: A Review-From Disease Metabolism to Precision Medicine.Biomed Res Int. 2019 Jun 9;2019:8304260. doi: 10.1155/2019/8304260. eCollection 2019. Biomed Res Int. 2019. PMID: 31281846 Free PMC article. Review.
-
Elucidating dynamic anaerobe metabolism with HRMAS 13C NMR and genome-scale modeling.Nat Chem Biol. 2023 May;19(5):556-564. doi: 10.1038/s41589-023-01275-9. Epub 2023 Mar 9. Nat Chem Biol. 2023. PMID: 36894723 Free PMC article.
References
-
- Janvilisri T, Scaria J, Thompson AD, Nicholson A, Limbago BM, Arroyo LG, Songer JG, Gröhn YT, Chang YF. Microarray identification of Clostridium difficile core components and divergent regions associated with host origin. J Bacteriol. 2009;191(12):3881–91. doi: 10.1128/JB.00222-09. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

