Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo
- PMID: 28209718
- PMCID: PMC5409871
- DOI: 10.1161/CIRCRESAHA.116.308741
Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo
Abstract
Rationale: Direct reprogramming of cardiac fibroblasts to cardiomyocytes has recently emerged as a novel and promising approach to regenerate the injured myocardium. We have previously demonstrated the feasibility of this approach in vitro and in vivo using a combination of 4 microRNAs (miR-1, miR-133, miR-208, and miR-499) that we named miR combo. However, the mechanism of miR combo mediated direct cardiac reprogramming is currently unknown.
Objective: Here, we investigated the possibility that miR combo initiated direct cardiac reprogramming through an epigenetic mechanism.
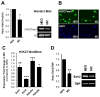
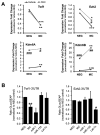
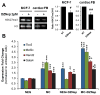
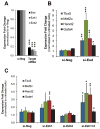
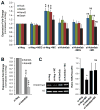
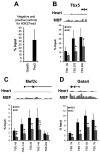
Methods and results: Using a quantitative polymerase chain reaction array, we found that histone methyltransferases and demethylases that regulate the trimethylation of H3K27 (H3K27me3), an epigenetic modification that marks transcriptional repression, were changed in miR combo-treated fibroblasts. Accordingly, global H3K27me3 levels were downregulated by miR combo treatment. In particular, the promoter region of cardiac transcription factors showed decreased H3K27me3 as revealed by chromatin immunoprecipitation coupled with quantitative polymerase chain reaction. Inhibition of H3K27 methyltransferases or of the PRC2 (Polycomb Repressive Complex 2) by pharmaceutical inhibition or siRNA reduced the levels of H3K27me3 and induced cardiogenic markers at the RNA and protein level, similarly to miR combo treatment. In contrast, knockdown of the H3K27 demethylases Kdm6A and Kdm6B restored the levels of H3K27me3 and blocked the induction of cardiac gene expression in miR combo-treated fibroblasts.
Conclusions: In summary, we demonstrated that removal of the repressive mark H3K27me3 is essential for the induction of cardiac reprogramming by miR combo. Our data not only highlight the importance of regulating the epigenetic landscape during cell fate conversion but also provide a framework to improve this technique.
Keywords: cardiac myocytes; cellular reprogramming; chromatin; epigenomics; microRNAs; regeneration.
© 2017 American Heart Association, Inc.
Figures
Comment in
-
Deciphering Histone Code Enigmas Sheds New Light on Cardiac Regeneration.Circ Res. 2017 Apr 28;120(9):1370-1372. doi: 10.1161/CIRCRESAHA.117.310919. Circ Res. 2017. PMID: 28450354 No abstract available.
References
-
- Dal-Pra S, Mirotsou M. Reprogramming approaches in cardiovascular regeneration. Curr Treat Options Cardiovasc Med. 2014;16:327. - PubMed
-
- Sadahiro T, Yamanaka S, Ieda M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ Res. 2015;116:1378–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

