The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality
- PMID: 28209721
- PMCID: PMC5399484
- DOI: 10.1182/blood-2016-10-748426
The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality
Abstract
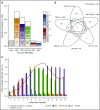
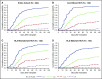
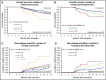
Strategies to prevent active infection with certain double-stranded DNA (dsDNA) viruses after allogeneic hematopoietic cell transplantation (HCT) are limited by incomplete understanding of their epidemiology and clinical impact. We retrospectively tested weekly plasma samples from allogeneic HCT recipients at our center from 2007 to 2014. We used quantitative PCR to test for cytomegalovirus, BK polyomavirus, human herpesvirus 6B, HHV-6A, adenovirus, and Epstein-Barr virus between days 0 and 100 post-HCT. We evaluated risk factors for detection of multiple viruses and association of viruses with mortality through day 365 post-HCT with Cox models. Among 404 allogeneic HCT recipients, including 125 cord blood, 125 HLA-mismatched, and 154 HLA-matched HCTs, detection of multiple viruses was common through day 100: 90% had ≥1, 62% had ≥2, 28% had ≥3, and 5% had 4 or 5 viruses. Risk factors for detection of multiple viruses included cord blood or HLA-mismatched HCT, myeloablative conditioning, and acute graft-versus-host disease (P values < .01). Absolute lymphocyte count of <200 cells/mm3 was associated with greater virus exposure on the basis of the maximum cumulative viral load area under the curve (AUC) (P = .054). The maximum cumulative viral load AUC was the best predictor of early (days 0-100) and late (days 101-365) overall mortality (adjusted hazard ratio [aHR] = 1.36, 95% confidence interval [CI] [1.25, 1.49], and aHR = 1.04, 95% CI [1.0, 1.08], respectively) after accounting for immune reconstitution and graft-versus-host disease. In conclusion, detection of multiple dsDNA viruses was frequent after allogeneic HCT and had a dose-dependent association with increased mortality. These data suggest opportunities to improve outcomes with better antiviral strategies.
© 2017 by The American Society of Hematology.
Figures


References
-
- Tomblyn M, Chiller T, Einsele H, et al. ; Center for International Blood and Marrow Research; National Marrow Donor program; European Blood and Marrow Transplant Group; American Society of Blood and Marrow Transplantation; Canadian Blood and Marrow Transplant Group; Infectious Diseases Society of America; Society for Healthcare Epidemiology of America; Association of Medical Microbiology and Infectious Disease Canada; Centers for Disease Control and Prevention. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant. 2009;15(10):1143-1238. - PMC - PubMed
-
- Kontoyiannis DP, Lewis RE, Marr K. The burden of bacterial and viral infections in hematopoietic stem cell transplant. Biol Blood Marrow Transplant. 2009;15(1 Suppl):128-133. - PubMed
-
- Mynarek M, Ganzenmueller T, Mueller-Heine A, et al. Patient, virus, and treatment-related risk factors in pediatric adenovirus infection after stem cell transplantation: results of a routine monitoring program. Biol Blood Marrow Transplant. 2014;20(2):250-256. - PubMed
-
- Rasche L, Kapp M, Einsele H, Mielke S. EBV-induced post transplant lymphoproliferative disorders: a persisting challenge in allogeneic hematopoetic SCT. Bone Marrow Transplant. 2014;49(2):163-167. - PubMed
-
- Rorije NMG, Shea MM, Satyanarayana G, et al. BK virus disease after allogeneic stem cell transplantation: a cohort analysis. Biol Blood Marrow Transplant. 2014;20(4):564-570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

