Distinctive Nuclear Localization Signals in the Oomycete Phytophthora sojae
- PMID: 28210240
- PMCID: PMC5288373
- DOI: 10.3389/fmicb.2017.00010
Distinctive Nuclear Localization Signals in the Oomycete Phytophthora sojae
Abstract
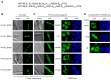
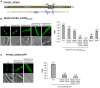
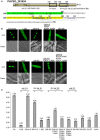
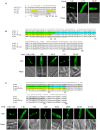
To date, nuclear localization signals (NLSs) that target proteins to nuclei in oomycetes have not been defined, but have been assumed to be the same as in higher eukaryotes. Here, we use the soybean pathogen Phytophthora sojae as a model to investigate these sequences in oomycetes. By establishing a reliable in vivo NLS assay based on confocal microscopy, we found that many canonical monopartite and bipartite classical NLSs (cNLSs) mediated nuclear import poorly in P. sojae. We found that efficient localization of P. sojae nuclear proteins by cNLSs requires additional basic amino acids at distal sites or collaboration with other NLSs. We found that several representatives of another well-characterized NLS, proline-tyrosine NLS (PY-NLS) also functioned poorly in P. sojae. To characterize PY-NLSs in P. sojae, we experimentally defined the residues required by functional PY-NLSs in three P. sojae nuclear-localized proteins. These results showed that functional P. sojae PY-NLSs include an additional cluster of basic residues for efficient nuclear import. Finally, analysis of several highly conserved P. sojae nuclear proteins including ribosomal proteins and core histones revealed that these proteins exhibit a similar but stronger set of sequence requirements for nuclear targeting compared with their orthologs in mammals or yeast.
Keywords: NLS; PY-NLS; Phytophthora sojae; classical NLS; nuclear localization; nucleus labeling; oomycetes.
Figures



References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B Methodol. 57, 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources

