The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon
- PMID: 28210761
- PMCID: PMC5744689
- DOI: 10.1007/s00239-017-9780-1
The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon
Abstract
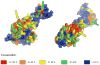
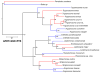
We screened the genomes of a broad panel of kinetoplastid protists for genes encoding proteins associated with the RNA interference (RNAi) system using probes from the Argonaute (AGO1), Dicer1 (DCL1), and Dicer2 (DCL2) genes of Leishmania brasiliensis and Crithidia fasciculata. We identified homologs for all the three of these genes in the genomes of a subset of these organisms. However, several of these organisms lacked evidence for any of these genes, while others lacked only DCL2. The open reading frames encoding these putative proteins were structurally analyzed in silico. The alignments indicated that the genes are homologous with a high degree of confidence, and three-dimensional structural models strongly supported a functional relationship to previously characterized AGO1, DCL1, and DCL2 proteins. Phylogenetic analysis of these putative proteins showed that these genes, when present, evolved in parallel with other nuclear genes, arguing that the RNAi system genes share a common progenitor, likely across all Kinetoplastea. In addition, the genome segments bearing these genes are highly conserved and syntenic, even among those taxa in which they are absent. However, taxa in which these genes are apparently absent represent several widely divergent branches of kinetoplastids, arguing that these genes were independently lost at least six times in the evolutionary history of these organisms. The mechanisms responsible for the apparent coordinate loss of these RNAi system genes independently in several lineages of kinetoplastids, while being maintained in other related lineages, are currently unknown.
Keywords: Evolutionary loss of genes; Kinetoplastea; RNAi; Trypanosomatida.
Figures


References
-
- Alves JMP, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, Teixeira MM, Motta MC, Camargo EP. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol. 2013a;13:190. - PMC - PubMed
-
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. - PMC - PubMed
-
- Barth S, Hury A, Liang XH, Michaeli S. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J Biol Chem. 2005;280:34558–34568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

