Multi-View Ensemble Classification of Brain Connectivity Images for Neurodegeneration Type Discrimination
- PMID: 28210983
- PMCID: PMC5443864
- DOI: 10.1007/s12021-017-9324-2
Multi-View Ensemble Classification of Brain Connectivity Images for Neurodegeneration Type Discrimination
Abstract
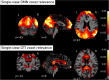
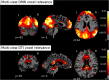
Brain connectivity analyses using voxels as features are not robust enough for single-patient classification because of the inter-subject anatomical and functional variability. To construct more robust features, voxels can be aggregated into clusters that are maximally coherent across subjects. Moreover, combining multi-modal neuroimaging and multi-view data integration techniques allows generating multiple independent connectivity features for the same patient. Structural and functional connectivity features were extracted from multi-modal MRI images with a clustering technique, and used for the multi-view classification of different phenotypes of neurodegeneration by an ensemble learning method (random forest). Two different multi-view models (intermediate and late data integration) were trained on, and tested for the classification of, individual whole-brain default-mode network (DMN) and fractional anisotropy (FA) maps, from 41 amyotrophic lateral sclerosis (ALS) patients, 37 Parkinson's disease (PD) patients and 43 healthy control (HC) subjects. Both multi-view data models exhibited ensemble classification accuracies significantly above chance. In ALS patients, multi-view models exhibited the best performances (intermediate: 82.9%, late: 80.5% correct classification) and were more discriminative than each single-view model. In PD patients and controls, multi-view models' performances were lower (PD: 59.5%, 62.2%; HC: 56.8%, 59.1%) but higher than at least one single-view model. Training the models only on patients, produced more than 85% patients correctly discriminated as ALS or PD type and maximal performances for multi-view models. These results highlight the potentials of mining complementary information from the integration of multiple data views in the classification of connectivity patterns from multi-modal brain images in the study of neurodegenerative diseases.
Keywords: Amyotrophic lateral sclerosis; Default mode network; Fractional anisotropy; Multi-modality; Multi-view; Parkinson’s disease; Random forests.
Conflict of interest statement
Conflict of Interest
The authors declare that they have no conflict of interest.
Figures






References
-
- Archer KJ, Kimes RV. Empirical characterization of random forest variable importance measures. Computational Statistics and Data Analysis. 2008;52:2249–2260. doi: 10.1016/j.csda.2007.08.015. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

