High-resolution 3D imaging of whole organ after clearing: taking a new look at the zebrafish testis
- PMID: 28211501
- PMCID: PMC5314416
- DOI: 10.1038/srep43012
High-resolution 3D imaging of whole organ after clearing: taking a new look at the zebrafish testis
Abstract
Zebrafish testis has become a powerful model for reproductive biology of teleostean fishes and other vertebrates and encompasses multiple applications in applied and basic research. Many studies have focused on 2D images, which is time consuming and implies extrapolation of results. Three-dimensional imaging of whole organs recently became an important challenge to better understand their architecture and allow cell enumeration. Several protocols have thus been developed to enhance sample transparency, a limiting step for imaging large biological samples. However, none of these methods has been applied to the zebrafish testis. We tested five clearing protocols to determine if some of them could be applied with only small modifications to the testis. We compared clearing efficiency at both macroscopic and microscopic levels. CUBIC and PACT were suitable for an efficient transparency, an optimal optical penetration, the GFP fluorescence preservation and avoiding meaningful tissue deformation. Finally, we succeeded in whole testis 3D capture at a cellular resolution with both CUBIC and PACT, which will be valuable in a standard workflow to investigate the 3D architecture of the testis and its cellular content. This paves the way for further development of high content phenotyping studies in several fields including development, genetic or toxicology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



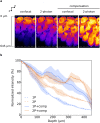

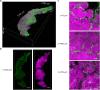

References
-
- Hama H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

