Expression profiling of TRIM protein family in THP1-derived macrophages following TLR stimulation
- PMID: 28211536
- PMCID: PMC5314404
- DOI: 10.1038/srep42781
Expression profiling of TRIM protein family in THP1-derived macrophages following TLR stimulation
Abstract
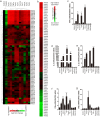
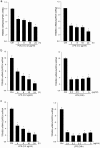
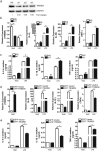
Activated macrophages play an important role in many inflammatory diseases including septic shock and atherosclerosis. However, the molecular mechanisms limiting macrophage activation are not completely understood. Members of the tripartite motif (TRIM) family have recently emerged as important players in innate immunity and antivirus. Here, we systematically analyzed mRNA expressions of representative TRIM molecules in human THP1-derived macrophages activated by different toll-like receptor (TLR) ligands. Twenty-nine TRIM members were highly induced (>3 fold) by one or more TLR ligands, among which 19 of them belong to TRIM C-IV subgroup. Besides TRIM21, TRIM22 and TRIM38 were shown to be upregulated by TLR3 and TLR4 ligands as previous reported, we identified a novel group of TRIM genes (TRIM14, 15, 31, 34, 43, 48, 49, 51 and 61) that were significantly up-regulated by TLR3 and TLR4 ligands. In contrast, the expression of TRIM59 was down-regulated by TLR3 and TLR4 ligands in both human and mouse macrophages. The alternations of the TRIM proteins were confirmed by Western blot. Finally, overexpression of TRIM59 significantly suppressed LPS-induced macrophage activation, whereas siRNA-mediated knockdown of TRIM59 enhanced LPS-induced macrophage activation. Taken together, the study provided an insight into the TLR ligands-induced expressions of TRIM family in macrophages.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Burke B., Sumner S., Maitland N. & Lewis C. E. Macrophages in gene therapy: cellular delivery vehicles and in vivo targets. Journal of leukocyte biology 72, 417–428 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

