SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication
- PMID: 28212444
- PMCID: PMC5333917
- DOI: 10.1371/journal.ppat.1006216
SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication
Abstract
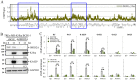
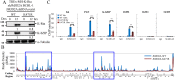
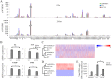
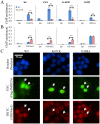
Small ubiquitin-like modifier (SUMO) modification of chromatin has profound effects on transcription regulation. By using Kaposi's sarcoma associated herpesvirus (KSHV) as a model, we recently demonstrated that epigenetic modification of viral chromatin by SUMO-2/3 is involved in regulating gene expression and viral reactivation. However, how this modification orchestrates transcription reprogramming through targeting histone modifying enzymes remains largely unknown. Here we show that JMJD2A, the first identified Jumonji C domain-containing histone demethylase, is the histone demethylase responsible for SUMO-2/3 enrichment on the KSHV genome during viral reactivation. Using in vitro and in vivo SUMOylation assays, we found that JMJD2A is SUMOylated on lysine 471 by KSHV K-bZIP, a viral SUMO-2/3-specific E3 ligase, in a SUMO-interacting motif (SIM)-dependent manner. SUMOylation is required for stabilizing chromatin association and gene transactivation by JMJD2A. These finding suggest that SUMO-2/3 modification plays an essential role in the epigenetic regulatory function of JMJD2A. Consistently, hierarchical clustering analysis of RNA-seq data showed that a SUMO-deficient mutant of JMJD2A was more closely related to JMJD2A knockdown than to wild-type. Our previous report demonstrated that JMJD2A coated and maintained the "ready to activate" status of the viral genome. Consistent with our previous report, a SUMO-deficient mutant of JMJD2A reduced viral gene expression and virion production. Importantly, JMJD2A has been implicated as an oncogene in various cancers by regulating proliferation. We therefore further analyzed the role of SUMO modification of JMJD2A in regulating cell proliferation. Interestingly, the SUMO-deficient mutant of JMJD2A failed to rescue the proliferation defect of JMJD2A knockdown cells. Emerging specific inhibitors of JMJD2A have been generated for evaluation in cancer studies. Our results revealed that SUMO conjugation mediates an epigenetic regulatory function of JMJD2A and suggests that inhibiting JMJD2A SUMOylation may be a novel avenue for anti-cancer therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

