Personalized medicine: Genetic risk prediction of drug response
- PMID: 28213088
- PMCID: PMC5653378
- DOI: 10.1016/j.pharmthera.2017.02.036
Personalized medicine: Genetic risk prediction of drug response
Erratum in
-
Corrigendum to "Personalized medicine: Genetic risk prediction of drug response" [Pharmacol. Ther. 175 (2017) 75-90.].Pharmacol Ther. 2018 Mar;183:205-206. doi: 10.1016/j.pharmthera.2017.12.001. Epub 2018 Jan 8. Pharmacol Ther. 2018. PMID: 29237545 No abstract available.
Abstract
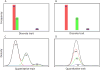
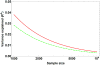
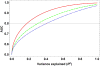
Pharmacogenomics (PGx), a substantial component of "personalized medicine", seeks to understand each individual's genetic composition to optimize drug therapy -- maximizing beneficial drug response, while minimizing adverse drug reactions (ADRs). Drug responses are highly variable because innumerable factors contribute to ultimate phenotypic outcomes. Recent genome-wide PGx studies have provided some insight into genetic basis of variability in drug response. These can be grouped into three categories. [a] Monogenic (Mendelian) traits include early examples mostly of inherited disorders, and some severe (idiosyncratic) ADRs typically influenced by single rare coding variants. [b] Predominantly oligogenic traits represent variation largely influenced by a small number of major pharmacokinetic or pharmacodynamic genes. [c] Complex PGx traits resemble most multifactorial quantitative traits -- influenced by numerous small-effect variants, together with epigenetic effects and environmental factors. Prediction of monogenic drug responses is relatively simple, involving detection of underlying mutations; due to rarity of these events and incomplete penetrance, however, prospective tests based on genotype will have high false-positive rates, plus pharmacoeconomics will require justification. Prediction of predominantly oligogenic traits is slowly improving. Although a substantial fraction of variation can be explained by limited numbers of large-effect genetic variants, uncertainty in successful predictions and overall cost-benefit ratios will make such tests elusive for everyday clinical use. Prediction of complex PGx traits is almost impossible in the foreseeable future. Genome-wide association studies of large cohorts will continue to discover relevant genetic variants; however, these small-effect variants, combined, explain only a small fraction of phenotypic variance -- thus having limited predictive power and clinical utility.
Keywords: Complex PGx traits; Genetic architecture; Genetic risk prediction; Monogenic (Mendelian) PGx traits; Pharmacogenetics; Pharmacogenomics (PGx); Predominantly oligogenetic PGx traits.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Both authors declare that they have no actual, or potential, conflicts of interest.
Figures
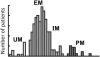

References
-
- Balwani M, Burrow TA, Charrow J, Goker-Alpan O, Kaplan P, Kishnani PS, Weinreb N. Recommendations for the use of eliglustat in the treatment of adults with Gaucher disease type 1 in the United States. Molecular Genetics and Metabolism. 2016;117:95–103. - PubMed
-
- Belmatoug N, Di Rocco M, Fraga C, Giraldo P, Hughes D, Lukina E, Cox TM. Management and monitoring recommendations for the use of eliglustat in adults with type 1 Gaucher disease in Europe. European Journal of Internal Medicine 2016 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

