Epimetabolites: discovering metabolism beyond building and burning
- PMID: 28213207
- PMCID: PMC5850962
- DOI: 10.1016/j.cbpa.2017.01.012
Epimetabolites: discovering metabolism beyond building and burning
Abstract
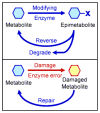
Enzymatic transformations of primary, canonical metabolites generate active biomolecules that regulate important cellular and physiological processes. Roles include regulation of histone demethylation in epigenetics, inflammation in tissue injury, insulin sensitivity, cancer cell invasion, stem cell pluripotency status, inhibition of nitric oxide signaling and others. Such modified compounds, defined as epimetabolites, have functions distinct from classic hormones as well as removed from generic anabolism and catabolism. Epimetabolites are discovered by untargeted metabolomics using liquid- or gas chromatography-high resolution mass spectrometry and structurally annotated by in-silico fragmentation prediction tools. Their specific biological functions are subsequently investigated by targeted metabolomics methods.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures


References
-
- O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural brain research. 2015;277:32–48. - PubMed
-
- Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nature chemical biology. 2013;9:72–80. - PubMed
-
- Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. The blood exposome and its role in discovering causes of disease. Environmental Health Perspectives (Online) 2014;122:769. This paper is a seminal contribution due to its comprehensive investigation of exposome compounds present in human blood, and ranges of concentrations that tightly overlap with levels of endogenous metabolites. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

