Timing the day: what makes bacterial clocks tick?
- PMID: 28216658
- PMCID: PMC5696799
- DOI: 10.1038/nrmicro.2016.196
Timing the day: what makes bacterial clocks tick?
Abstract
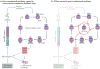
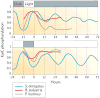
Chronobiological studies of prokaryotic organisms have generally lagged far behind the study of endogenous circadian clocks in eukaryotes, in which such systems are essentially ubiquitous. However, despite only being studied during the past 25 years, cyanobacteria have become important model organisms for the study of circadian rhythms and, presently, their timekeeping mechanism is the best understood of any system in terms of biochemistry, structural biology, biophysics and adaptive importance. Nevertheless, intrinsic daily rhythmicity among bacteria other than cyanobacteria is essentially unknown; some tantalizing information suggests widespread daily timekeeping among Eubacteria and Archaea through mechanisms that share common elements with the cyanobacterial clock but are distinct. Moreover, the recent surge of information about microbiome-host interactions has largely neglected the temporal dimension and yet daily cycles control important aspects of their relationship.
Figures




References
-
- Grobbelaar N, Huang T, Lin H, Chow T. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol Lett. 1986;37:173–177. This is the first persuasive report of a circadian rhythm expressed by a bacterium.
-
- Kondo T, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. This paper establishes the cyanobacterial circadian system that has been so productive in understanding the mechanism and adaptive importance of circadian rhythms. - PMC - PubMed
-
- Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. - PubMed
-
- Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

