Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics
- PMID: 28217095
- PMCID: PMC5290407
- DOI: 10.3389/fphar.2017.00023
Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics
Abstract
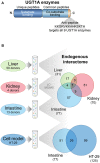
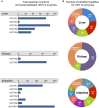
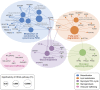
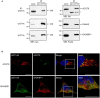
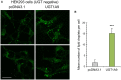
The conjugative metabolism mediated by UDP-glucuronosyltransferase enzymes (UGTs) significantly influences the bioavailability and biological responses of endogenous molecule substrates and xenobiotics including drugs. UGTs participate in the regulation of cellular homeostasis by limiting stress induced by toxic molecules, and by controlling hormonal signaling networks. Glucuronidation is highly regulated at genomic, transcriptional, post-transcriptional and post-translational levels. However, the UGT protein interaction network, which is likely to influence glucuronidation, has received little attention. We investigated the endogenous protein interactome of human UGT1A enzymes in main drug metabolizing non-malignant tissues where UGT expression is most prevalent, using an unbiased proteomics approach. Mass spectrometry analysis of affinity-purified UGT1A enzymes and associated protein complexes in liver, kidney and intestine tissues revealed an intricate interactome linking UGT1A enzymes to multiple metabolic pathways. Several proteins of pharmacological importance such as transferases (including UGT2 enzymes), transporters and dehydrogenases were identified, upholding a potential coordinated cellular response to small lipophilic molecules and drugs. Furthermore, a significant cluster of functionally related enzymes involved in fatty acid β-oxidation, as well as in the glycolysis and glycogenolysis pathways were enriched in UGT1A enzymes complexes. Several partnerships were confirmed by co-immunoprecipitations and co-localization by confocal microscopy. An enhanced accumulation of lipid droplets in a kidney cell model overexpressing the UGT1A9 enzyme supported the presence of a functional interplay. Our work provides unprecedented evidence for a functional interaction between glucuronidation and bioenergetic metabolism.
Keywords: UGT; affinity purification; human tissues; mass spectrometry; metabolism; protein-protein interaction; proteomics.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

