Genomic Characterization of Dairy Associated Leuconostoc Species and Diversity of Leuconostocs in Undefined Mixed Mesophilic Starter Cultures
- PMID: 28217118
- PMCID: PMC5289962
- DOI: 10.3389/fmicb.2017.00132
Genomic Characterization of Dairy Associated Leuconostoc Species and Diversity of Leuconostocs in Undefined Mixed Mesophilic Starter Cultures
Abstract
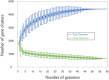
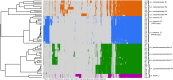
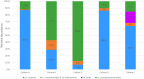
Undefined mesophilic mixed (DL-type) starter cultures are composed of predominantly Lactococcus lactis subspecies and 1-10% Leuconostoc spp. The composition of the Leuconostoc population in the starter culture ultimately affects the characteristics and the quality of the final product. The scientific basis for the taxonomy of dairy relevant leuconostocs can be traced back 50 years, and no documentation on the genomic diversity of leuconostocs in starter cultures exists. We present data on the Leuconostoc population in five DL-type starter cultures commonly used by the dairy industry. The analyses were performed using traditional cultivation methods, and further augmented by next-generation DNA sequencing methods. Bacterial counts for starter cultures cultivated on two different media, MRS and MPCA, revealed large differences in the relative abundance of leuconostocs. Most of the leuconostocs in two of the starter cultures were unable to grow on MRS, emphasizing the limitations of culture-based methods and the importance of careful media selection or use of culture independent methods. Pan-genomic analysis of 59 Leuconostoc genomes enabled differentiation into twelve robust lineages. The genomic analyses show that the dairy-associated leuconostocs are highly adapted to their environment, characterized by the acquisition of genotype traits, such as the ability to metabolize citrate. In particular, Leuconostoc mesenteroides subsp. cremoris display telltale signs of a degenerative evolution, likely resulting from a long period of growth in milk in association with lactococci. Great differences in the metabolic potential between Leuconostoc species and subspecies were revealed. Using targeted amplicon sequencing, the composition of the Leuconostoc population in the five commercial starter cultures was shown to be significantly different. Three of the cultures were dominated by Ln. mesenteroides subspecies cremoris. Leuconostoc pseudomesenteroides dominated in two of the cultures while Leuconostoc lactis, reported to be a major constituent in fermented dairy products, was only present in low amounts in one of the cultures. This is the first in-depth study of Leuconostoc genomics and diversity in dairy starter cultures. The results and the techniques presented may be of great value for the dairy industry.
Keywords: cheese; comparative; dairy; differentiation; diversity analysis; genomics; leuconostoc; starter cultures.
Figures

Similar articles
-
Lactococcus lactis Diversity in Undefined Mixed Dairy Starter Cultures as Revealed by Comparative Genome Analyses and Targeted Amplicon Sequencing of epsD.Appl Environ Microbiol. 2018 Jan 17;84(3):e02199-17. doi: 10.1128/AEM.02199-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29222100 Free PMC article.
-
Metagenomic Analysis of Dairy Bacteriophages: Extraction Method and Pilot Study on Whey Samples Derived from Using Undefined and Defined Mesophilic Starter Cultures.Appl Environ Microbiol. 2017 Sep 15;83(19):e00888-17. doi: 10.1128/AEM.00888-17. Print 2017 Oct 1. Appl Environ Microbiol. 2017. PMID: 28754704 Free PMC article.
-
Characterization of starter lactic acid bacteria from the Finnish fermented milk product viili.J Appl Microbiol. 2008 Dec;105(6):1929-38. doi: 10.1111/j.1365-2672.2008.03952.x. J Appl Microbiol. 2008. PMID: 19120639
-
The use of mesophilic cultures in the dairy industry.Antonie Van Leeuwenhoek. 1983 Sep;49(3):297-312. doi: 10.1007/BF00399505. Antonie Van Leeuwenhoek. 1983. PMID: 6414367 Review.
-
Bacteriocins produced by Leuconostoc species.J Dairy Sci. 1994 Sep;77(9):2718-24. doi: 10.3168/jds.S0022-0302(94)77214-3. J Dairy Sci. 1994. PMID: 7814741 Review.
Cited by
-
Genetic Diversity of Leuconostoc mesenteroides Isolates from Traditional Montenegrin Brine Cheese.Microorganisms. 2021 Jul 28;9(8):1612. doi: 10.3390/microorganisms9081612. Microorganisms. 2021. PMID: 34442691 Free PMC article.
-
Antilisterial activity of raw sheep milk from two native Epirus breeds: Culture-dependent identification, bacteriocin gene detection and primary safety evaluation of the antagonistic LAB biota.Curr Res Microb Sci. 2023 Dec 10;6:100209. doi: 10.1016/j.crmicr.2023.100209. eCollection 2024. Curr Res Microb Sci. 2023. PMID: 38116185 Free PMC article.
-
ODFM, an omics data resource from microorganisms associated with fermented foods.Sci Data. 2021 Apr 20;8(1):113. doi: 10.1038/s41597-021-00895-x. Sci Data. 2021. PMID: 33879798 Free PMC article.
-
Draft genome sequence of Leuconostoc falkenbergense isolated from naturally fermented buffalo milk curd.Microbiol Resour Announc. 2024 Apr 11;13(5):e0014824. doi: 10.1128/mra.00148-24. Online ahead of print. Microbiol Resour Announc. 2024. PMID: 38602401 Free PMC article.
-
Comparative genomics of Loigolactobacillus coryniformis with an emphasis on L. coryniformis strain FOL-19 isolated from cheese.Comput Struct Biotechnol J. 2023 Oct 14;21:5111-5124. doi: 10.1016/j.csbj.2023.10.004. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37920811 Free PMC article.
References
-
- Alegria A., Delgado S., Florez A. B., Mayo B. (2013). Identification, typing, and functional characterization of Leuconostoc spp. strains from traditional, starter-free cheeses. Dairy Sci. Technol. 93, 657–673. 10.1007/s13594-013-0128-3 - DOI
-
- Ardö Y., Varming C. (2010). Bacterial influence on characteristic flavour of cheeses made with mesophilic DL-starter. Aust. J. Dairy Technol. 65, 153–158.
-
- Auty M. A., Gardiner G. E., McBrearty S. J., O'Sullivan E. O., Mulvihill D. M., Collins J. K., et al. . (2001). Direct in situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl. Environ. Microbiol. 67, 420–425. 10.1128/AEM.67.1.420-425.2001 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

