Regulation of Antimicrobial Peptides in Aedes aegypti Aag2 Cells
- PMID: 28217557
- PMCID: PMC5291090
- DOI: 10.3389/fcimb.2017.00022
Regulation of Antimicrobial Peptides in Aedes aegypti Aag2 Cells
Abstract
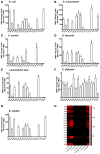
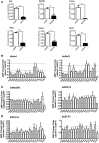
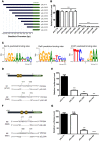
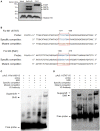
Antimicrobial peptides (AMPs) are an important group of immune effectors that play a role in combating microbial infections in invertebrates. Most of the current information on the regulation of insect AMPs in microbial infection have been gained from Drosophila, and their regulation in other insects are still not completely understood. Here, we generated an AMP induction profile in response to infections with some Gram-negative, -positive bacteria, and fungi in Aedes aegypti embryonic Aag2 cells. Most of the AMP inductions caused by the gram-negative bacteria was controlled by the Immune deficiency (Imd) pathway; nonetheless, Gambicin, an AMP gene discovered only in mosquitoes, was combinatorially regulated by the Imd, Toll and JAK-STAT pathways in the Aag2 cells. Gambicin promoter analyses including specific sequence motif deletions implicated these three pathways in Gambicin activity, as shown by a luciferase assay. Moreover, the recognition between Rel1 (refer to Dif/Dorsal in Drosophila) and STAT and their regulatory sites at the Gambicin promoter site was validated by a super-shift electrophoretic mobility shift assay (EMSA). Our study provides information that increases our understanding of the regulation of AMPs in response to microbial infections in mosquitoes. And it is a new finding that the A. aegypti AMPs are mainly regulated Imd pathway only, which is quite different from the previous understanding obtained from Drosophila.
Keywords: antimicrobial peptides; innate immunity; insect immunity; mosquito; regulation.
Figures
Similar articles
-
The Aedes aegypti IMD pathway is a critical component of the mosquito antifungal immune response.Dev Comp Immunol. 2019 Jun;95:1-9. doi: 10.1016/j.dci.2018.12.010. Epub 2018 Dec 22. Dev Comp Immunol. 2019. PMID: 30582948
-
Dengue virus inhibits immune responses in Aedes aegypti cells.PLoS One. 2010 May 18;5(5):e10678. doi: 10.1371/journal.pone.0010678. PLoS One. 2010. PMID: 20502529 Free PMC article.
-
Functional characterization of two clip domain serine proteases in innate immune responses of Aedes aegypti.Parasit Vectors. 2021 Nov 24;14(1):584. doi: 10.1186/s13071-021-05091-9. Parasit Vectors. 2021. PMID: 34819136 Free PMC article.
-
Defensive remodeling: How bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides.Biochim Biophys Acta. 2015 Nov;1848(11 Pt B):3089-100. doi: 10.1016/j.bbamem.2015.05.022. Epub 2015 Jun 4. Biochim Biophys Acta. 2015. PMID: 26051126 Review.
-
Resistance to antimicrobial peptides in Gram-negative bacteria.FEMS Microbiol Lett. 2012 May;330(2):81-9. doi: 10.1111/j.1574-6968.2012.02528.x. Epub 2012 Mar 12. FEMS Microbiol Lett. 2012. PMID: 22339775 Review.
Cited by
-
Molecular Rationale of Insect-Microbes Symbiosis-From Insect Behaviour to Mechanism.Microorganisms. 2021 Nov 24;9(12):2422. doi: 10.3390/microorganisms9122422. Microorganisms. 2021. PMID: 34946024 Free PMC article. Review.
-
Mosquito antiviral defense mechanisms: a delicate balance between innate immunity and persistent viral infection.Parasit Vectors. 2019 Apr 11;12(1):165. doi: 10.1186/s13071-019-3433-8. Parasit Vectors. 2019. PMID: 30975197 Free PMC article. Review.
-
Carry-over effects of Bacillus thuringiensis on tolerant Aedes albopictus mosquitoes.Parasit Vectors. 2024 Nov 7;17(1):456. doi: 10.1186/s13071-024-06556-3. Parasit Vectors. 2024. PMID: 39511654 Free PMC article.
-
Interspecies Isobaric Labeling-Based Quantitative Proteomics Reveals Protein Changes in the Ovary of Aedes aegypti Coinfected With ZIKV and Wolbachia.Front Cell Infect Microbiol. 2022 Jul 7;12:900608. doi: 10.3389/fcimb.2022.900608. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35873163 Free PMC article.
-
Dengue Virus-2 Infection Affects Fecundity and Elicits Specific Transcriptional Changes in the Ovaries of Aedes aegypti Mosquitoes.Front Microbiol. 2022 Jun 23;13:886787. doi: 10.3389/fmicb.2022.886787. eCollection 2022. Front Microbiol. 2022. PMID: 35814655 Free PMC article.
References
-
- Bulet P., Dimarcq J. L., Hetru C., Lagueux M., Charlet M., Hegy G., et al. . (1993). A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 268, 14893–14897. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

