Pneumococcal prophages are diverse, but not without structure or history
- PMID: 28218261
- PMCID: PMC5317160
- DOI: 10.1038/srep42976
Pneumococcal prophages are diverse, but not without structure or history
Abstract
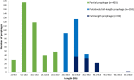
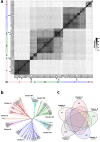
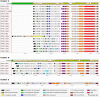
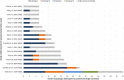
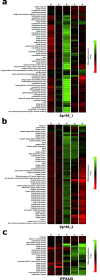
Bacteriophages (phages) infect many bacterial species, but little is known about the diversity of phages among the pneumococcus, a leading global pathogen. The objectives of this study were to determine the prevalence, diversity and molecular epidemiology of prophages (phage DNA integrated within the bacterial genome) among pneumococci isolated over the past 90 years. Nearly 500 pneumococcal genomes were investigated and RNA sequencing was used to explore prophage gene expression. We revealed that every pneumococcal genome contained prophage DNA. 286 full-length/putatively full-length pneumococcal prophages were identified, of which 163 have not previously been reported. Full-length prophages clustered into four major groups and every group dated from the 1930-40 s onward. There was limited evidence for genes shared between prophage clusters. Prophages typically integrated in one of five different sites within the pneumococcal genome. 72% of prophages possessed the virulence genes pblA and/or pblB. Individual prophages and the host pneumococcal genetic lineage were strongly associated and some prophages persisted for many decades. RNA sequencing provided clear evidence of prophage gene expression. Overall, pneumococcal prophages were highly prevalent, demonstrated a structured population, possessed genes associated with virulence, and were expressed under experimental conditions. Pneumococcal prophages are likely to play a more important role in pneumococcal biology and evolution than previously recognised.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Anthony D. & Mullerbeck E. Committing to child survival: a promise renewed. United Nations Children’s Fund (UNICEF) (2013).
-
- Liu L. et al. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385, 430–440 (2015). - PubMed
-
- World Health Organization. Pneumococcal vaccines WHO position paper – 2012. Wkly. Epidemiol. Rec. 87, 129–144 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

