Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks
- PMID: 28218679
- PMCID: PMC5333062
- DOI: 10.3390/genes8020073
Mcm10: A Dynamic Scaffold at Eukaryotic Replication Forks
Abstract
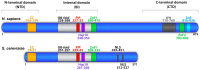
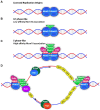
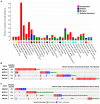
To complete the duplication of large genomes efficiently, mechanisms have evolved that coordinate DNA unwinding with DNA synthesis and provide quality control measures prior to cell division. Minichromosome maintenance protein 10 (Mcm10) is a conserved component of the eukaryotic replisome that contributes to this process in multiple ways. Mcm10 promotes the initiation of DNA replication through direct interactions with the cell division cycle 45 (Cdc45)-minichromosome maintenance complex proteins 2-7 (Mcm2-7)-go-ichi-ni-san GINS complex proteins, as well as single- and double-stranded DNA. After origin firing, Mcm10 controls replication fork stability to support elongation, primarily facilitating Okazaki fragment synthesis through recruitment of DNA polymerase-α and proliferating cell nuclear antigen. Based on its multivalent properties, Mcm10 serves as an essential scaffold to promote DNA replication and guard against replication stress. Under pathological conditions, Mcm10 is often dysregulated. Genetic amplification and/or overexpression of MCM10 are common in cancer, and can serve as a strong prognostic marker of poor survival. These findings are compatible with a heightened requirement for Mcm10 in transformed cells to overcome limitations for DNA replication dictated by altered cell cycle control. In this review, we highlight advances in our understanding of when, where and how Mcm10 functions within the replisome to protect against barriers that cause incomplete replication.
Keywords: CMG helicase; DNA replication; Mcm10; genome stability; origin activation; replication elongation; replication initiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

