The genome landscape of indigenous African cattle
- PMID: 28219390
- PMCID: PMC5319050
- DOI: 10.1186/s13059-017-1153-y
The genome landscape of indigenous African cattle
Abstract
Background: The history of African indigenous cattle and their adaptation to environmental and human selection pressure is at the root of their remarkable diversity. Characterization of this diversity is an essential step towards understanding the genomic basis of productivity and adaptation to survival under African farming systems.
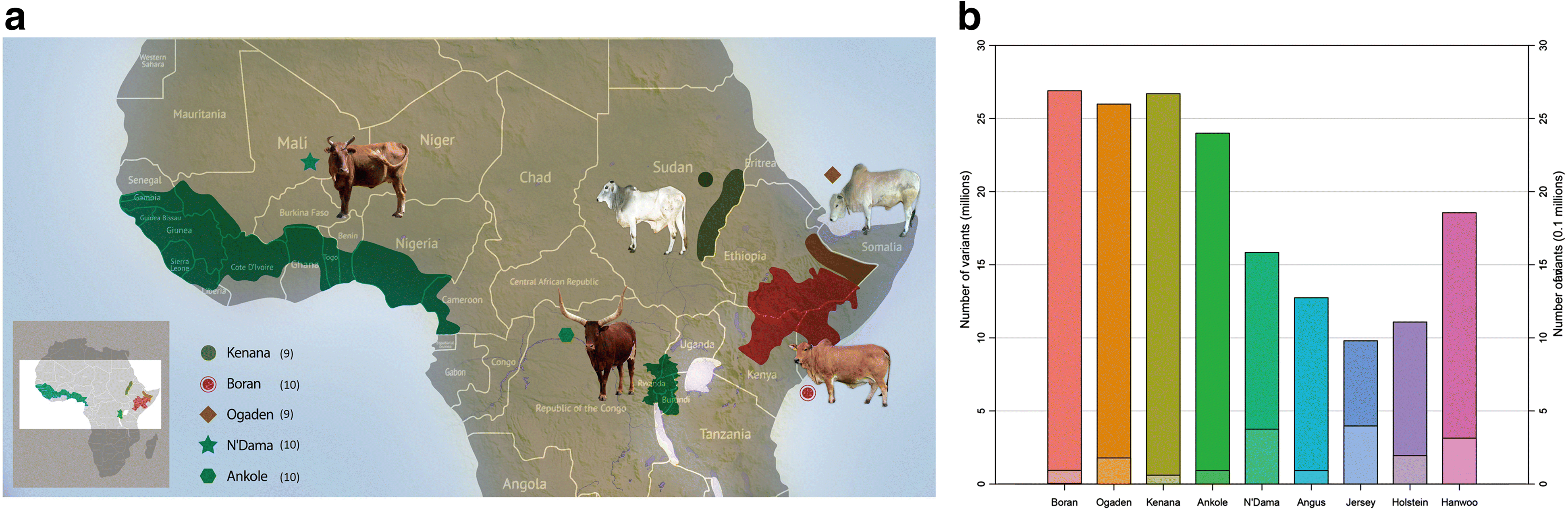
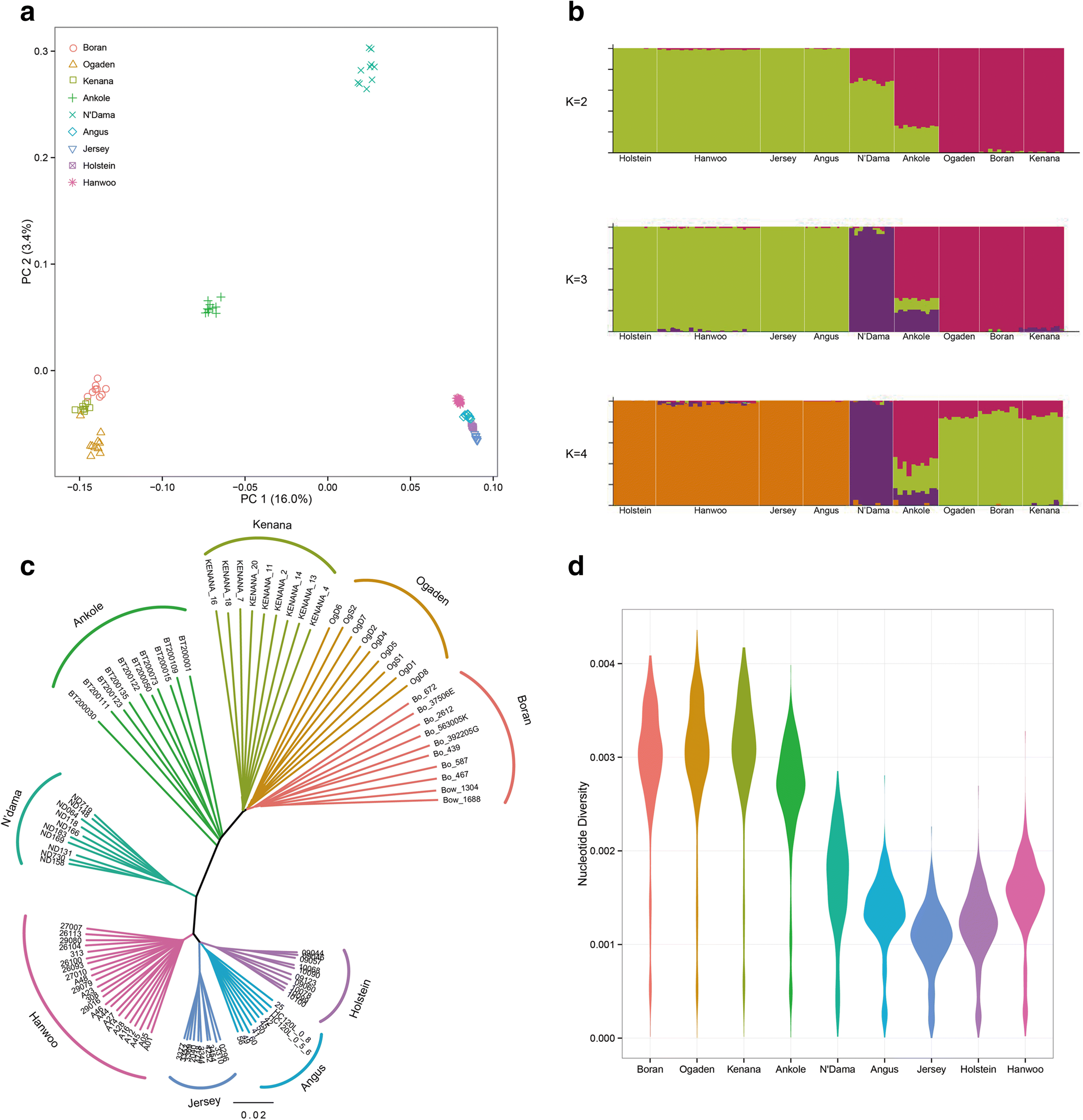
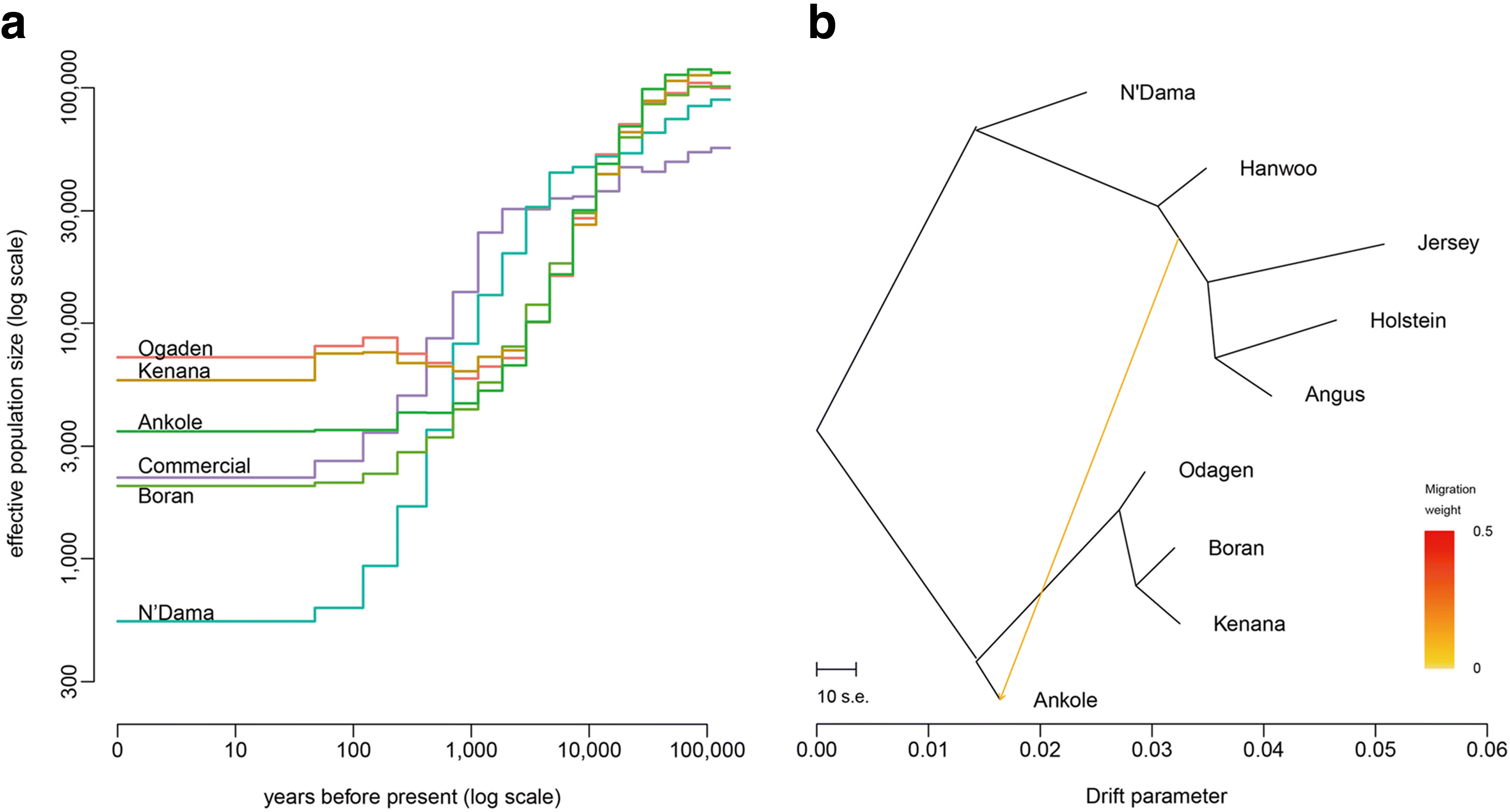
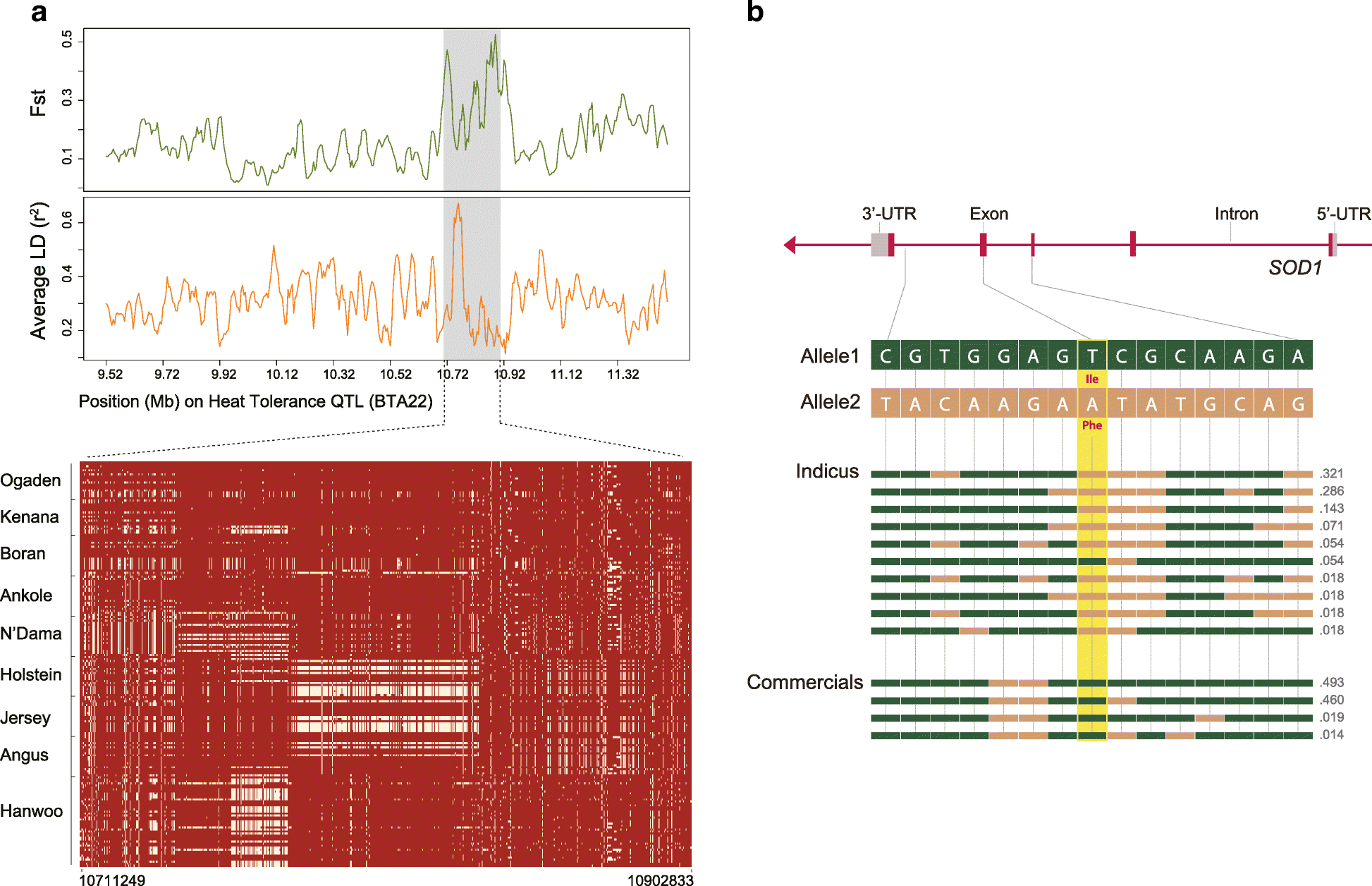
Results: We analyze patterns of African cattle genetic variation by sequencing 48 genomes from five indigenous populations and comparing them to the genomes of 53 commercial taurine breeds. We find the highest genetic diversity among African zebu and sanga cattle. Our search for genomic regions under selection reveals signatures of selection for environmental adaptive traits. In particular, we identify signatures of selection including genes and/or pathways controlling anemia and feeding behavior in the trypanotolerant N'Dama, coat color and horn development in Ankole, and heat tolerance and tick resistance across African cattle especially in zebu breeds.
Conclusions: Our findings unravel at the genome-wide level, the unique adaptive diversity of African cattle while emphasizing the opportunities for sustainable improvement of livestock productivity on the continent.
Keywords: Adaptation; African cattle; Diversity; Genome.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

