RNAs competing for microRNAs mutually influence their fluctuations in a highly non-linear microRNA-dependent manner in single cells
- PMID: 28219439
- PMCID: PMC5319025
- DOI: 10.1186/s13059-017-1162-x
RNAs competing for microRNAs mutually influence their fluctuations in a highly non-linear microRNA-dependent manner in single cells
Abstract
Background: Distinct RNA species may compete for binding to microRNAs (miRNAs). This competition creates an indirect interaction between miRNA targets, which behave as miRNA sponges and eventually influence each other's expression levels. Theoretical predictions suggest that not only the mean expression levels of targets but also the fluctuations around the means are coupled through miRNAs. This may result in striking effects on a broad range of cellular processes, such as cell differentiation and proliferation. Although several studies have reported the functional relevance of this mechanism of interaction, detailed experiments are lacking that study this phenomenon in controlled conditions by mimicking a physiological range.
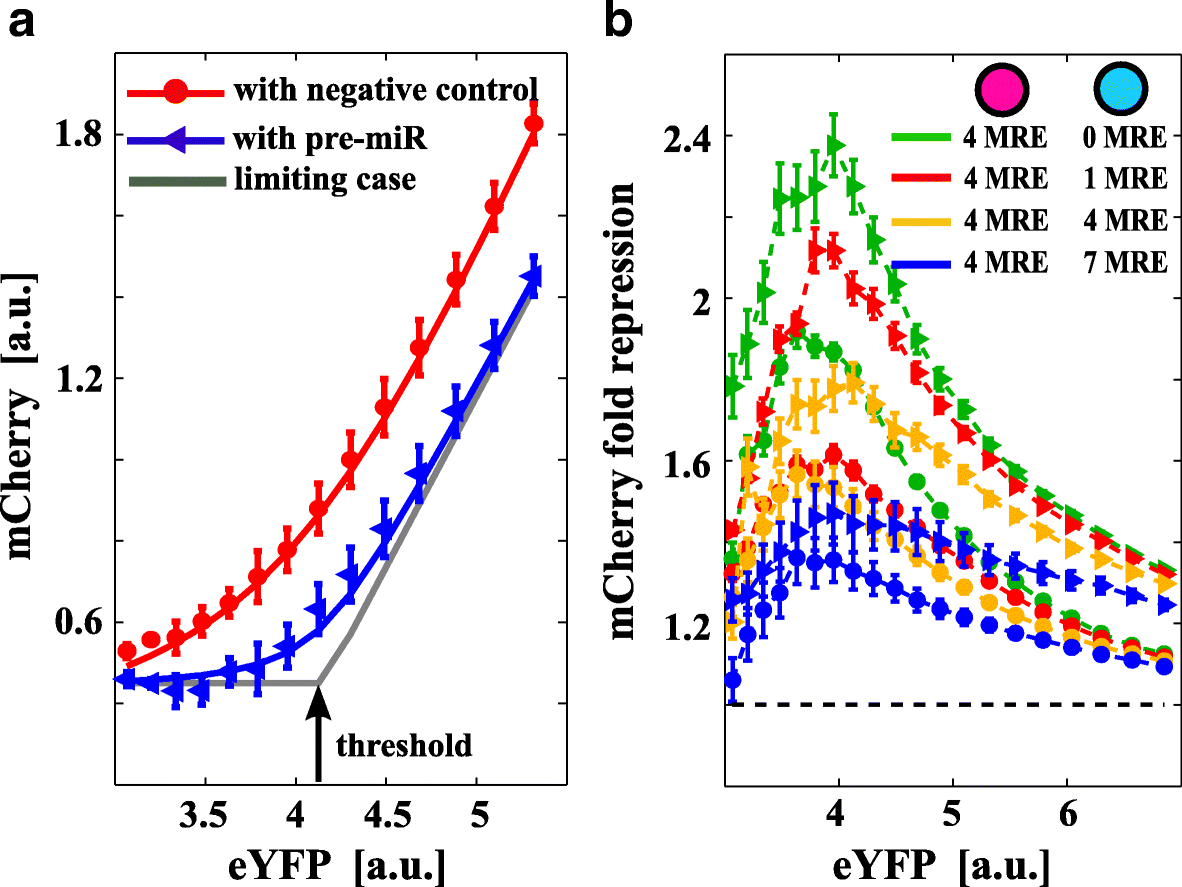
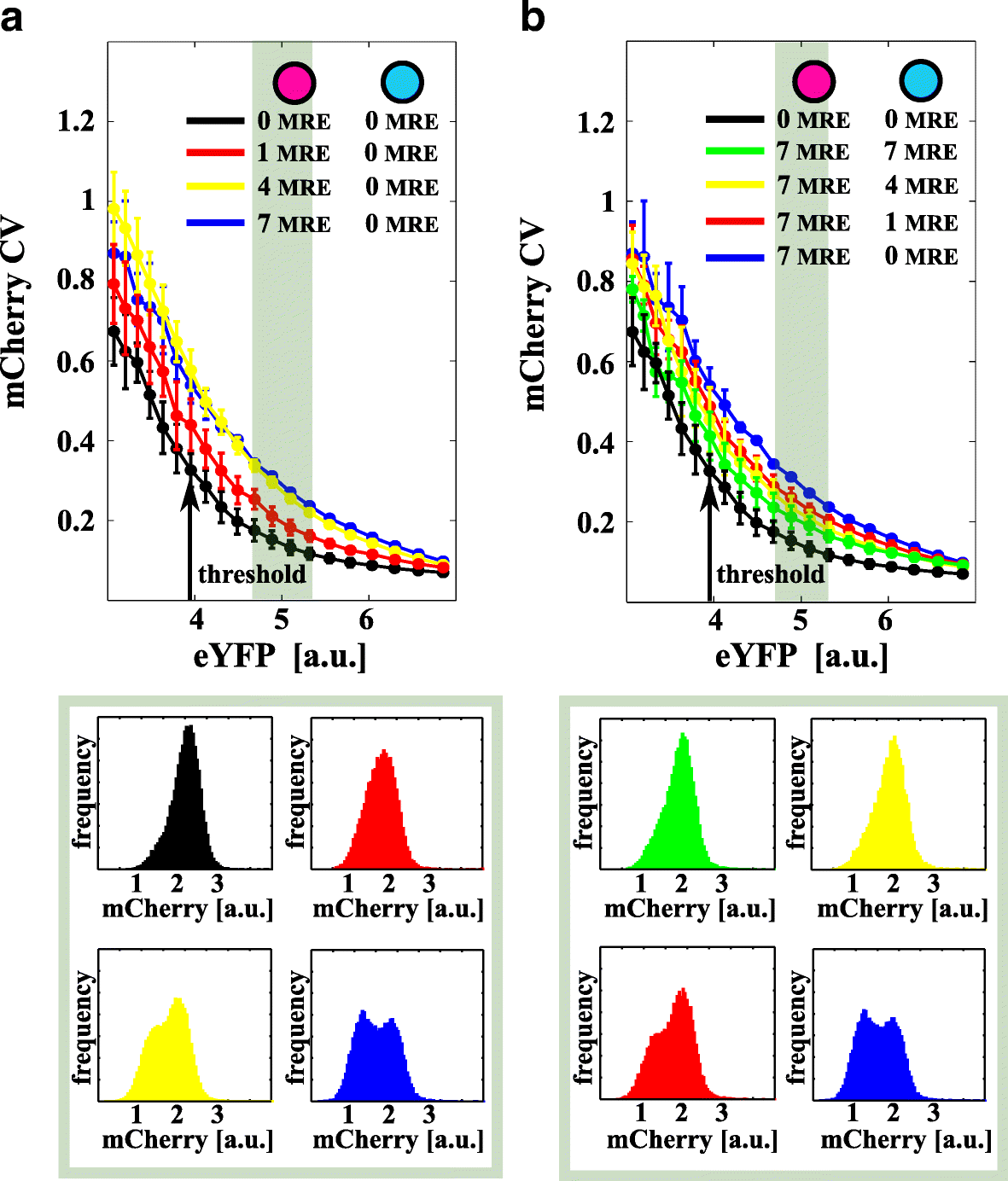
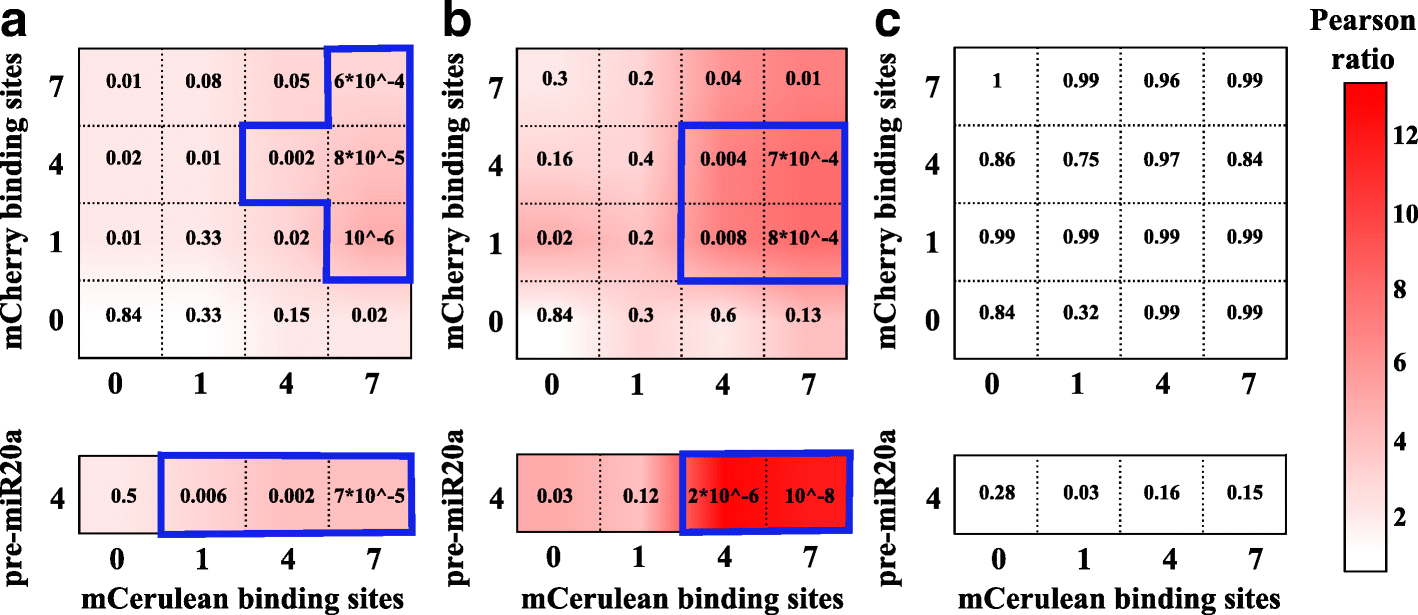
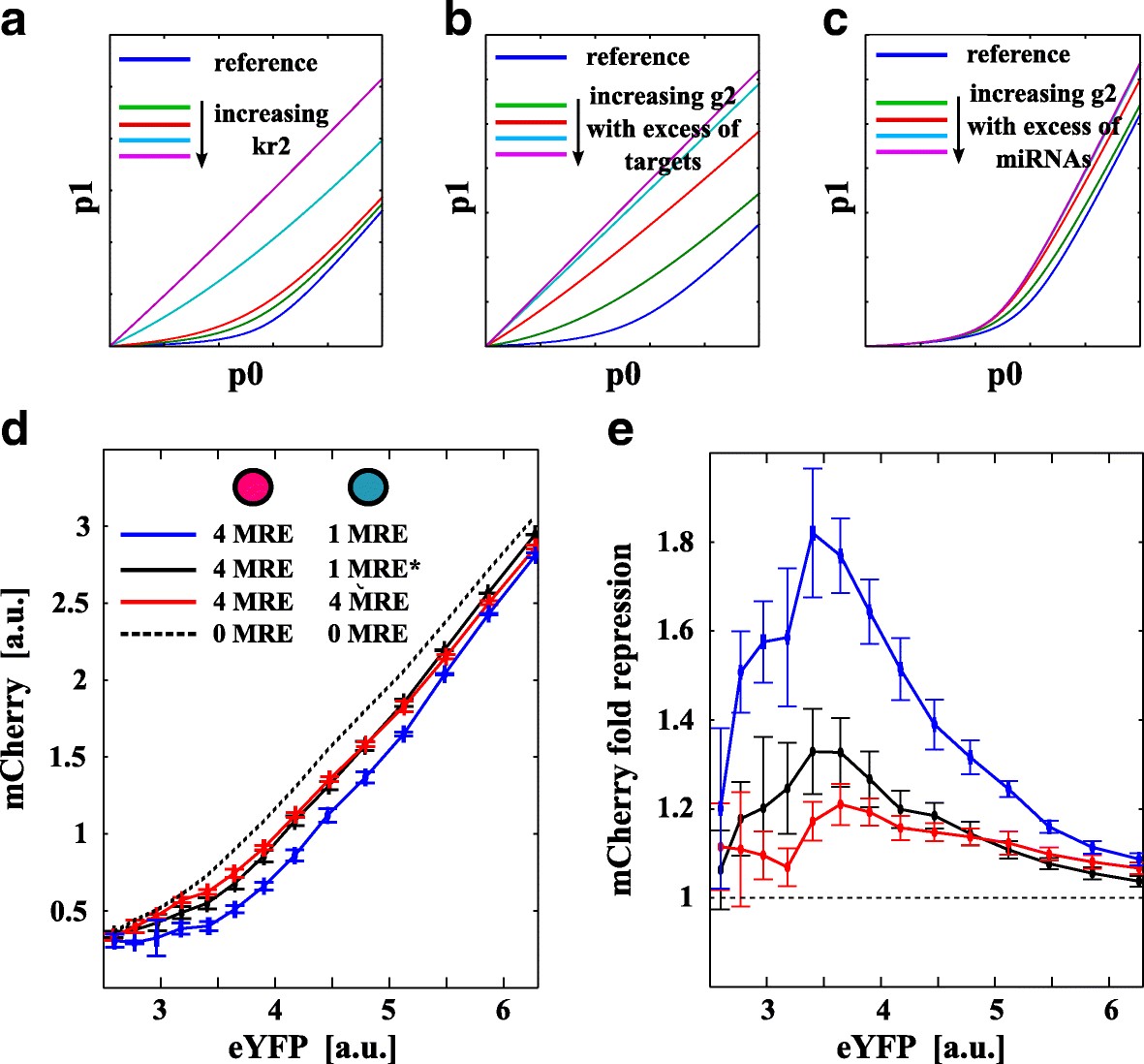
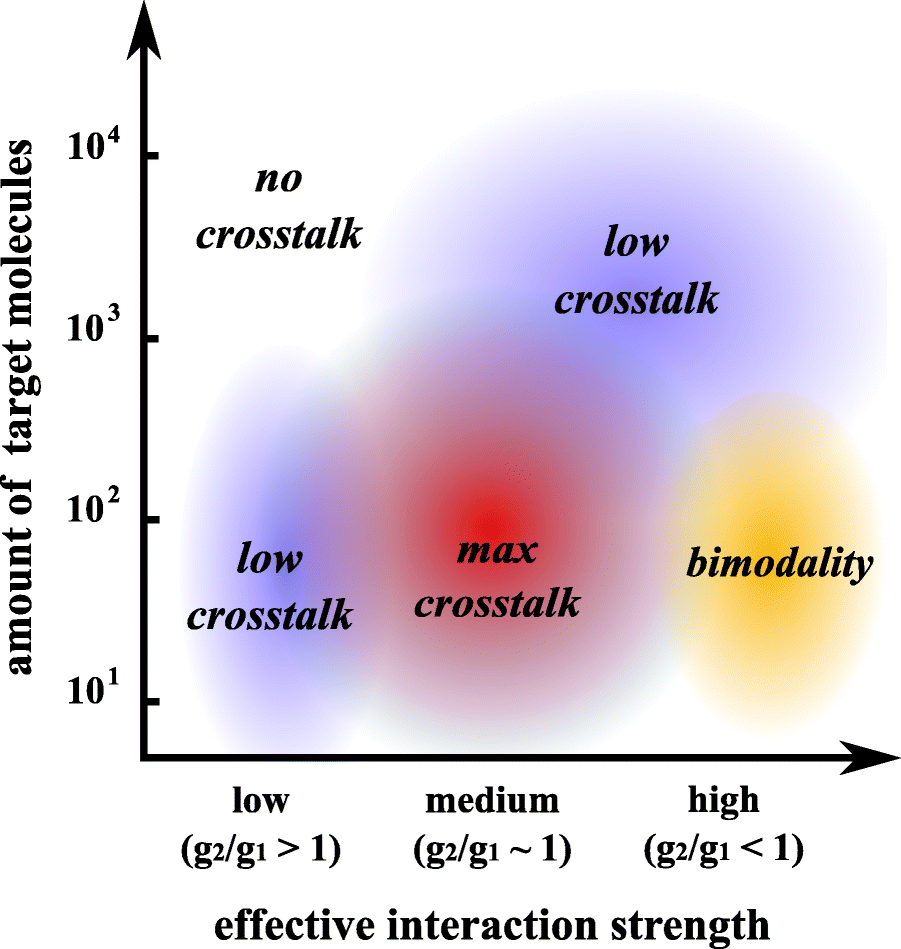
Results: We used an experimental design based on two bidirectional plasmids and flow cytometry measurements of cotransfected mammalian cells. We validated a stochastic gene interaction model that describes how mRNAs can influence each other's fluctuations in a miRNA-dependent manner in single cells. We show that miRNA-target correlations eventually lead to either bimodal cell population distributions with high and low target expression states, or correlated fluctuations across targets when the pool of unbound targets and miRNAs are in near-equimolar concentration. We found that there is an optimal range of conditions for the onset of cross-regulation, which is compatible with 10-1000 copies of targets per cell.
Conclusions: Our results are summarized in a phase diagram for miRNA-mediated cross-regulation that links experimentally measured quantities and effective model parameters. This phase diagram can be applied to in vivo studies of RNAs that are in competition for miRNA binding.
Keywords: Bimodality; MicroRNA target synchronization; Post-transcriptional cross-regulation; Single cell; Stochastic modelling.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

