A functional SNP associated with atopic dermatitis controls cell type-specific methylation of the VSTM1 gene locus
- PMID: 28219444
- PMCID: PMC5319034
- DOI: 10.1186/s13073-017-0404-6
A functional SNP associated with atopic dermatitis controls cell type-specific methylation of the VSTM1 gene locus
Abstract
Background: Expression quantitative trait loci (eQTL) databases represent a valuable resource to link disease-associated SNPs to specific candidate genes whose gene expression is significantly modulated by the SNP under investigation. We previously identified signal inhibitory receptor on leukocytes-1 (SIRL-1) as a powerful regulator of human innate immune cell function. While it is constitutively high expressed on neutrophils, on monocytes the SIRL-1 surface expression varies strongly between individuals. The underlying mechanism of regulation, its genetic control as well as potential clinical implications had not been explored yet.
Methods: Whole blood eQTL data of a Chinese cohort was used to identify SNPs regulating the expression of VSTM1, the gene encoding SIRL-1. The genotype effect was validated by flow cytometry (cell surface expression), correlated with electrophoretic mobility shift assay (EMSA), chromatin immunoprecipitation (ChIP) and bisulfite sequencing (C-methylation) and its functional impact studied the inhibition of reactive oxygen species (ROS).
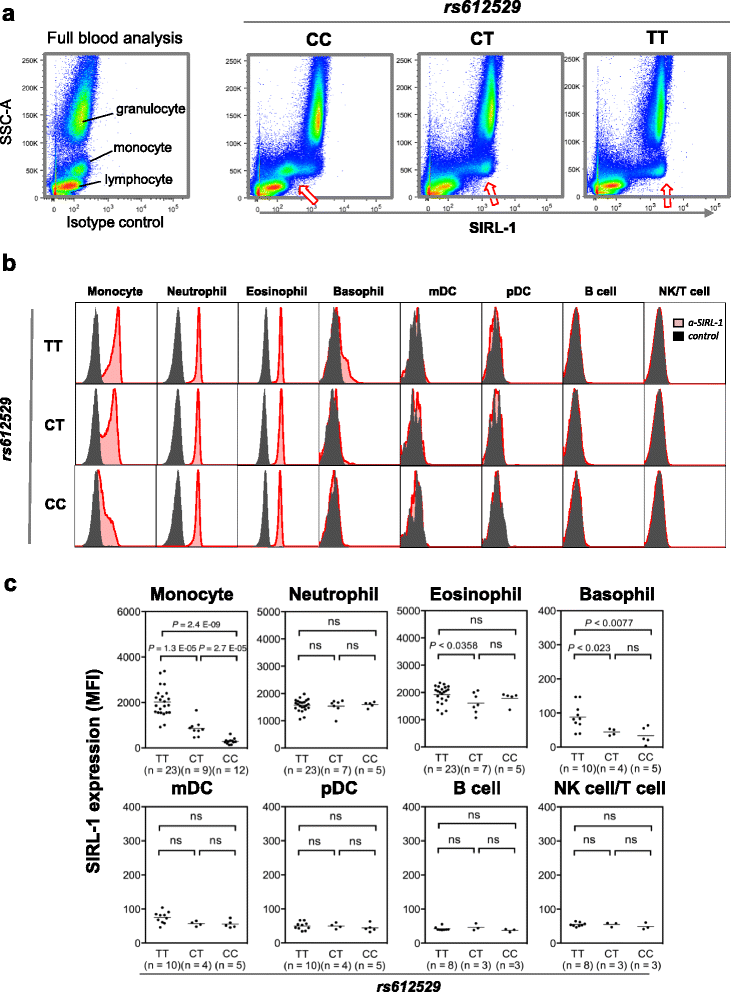
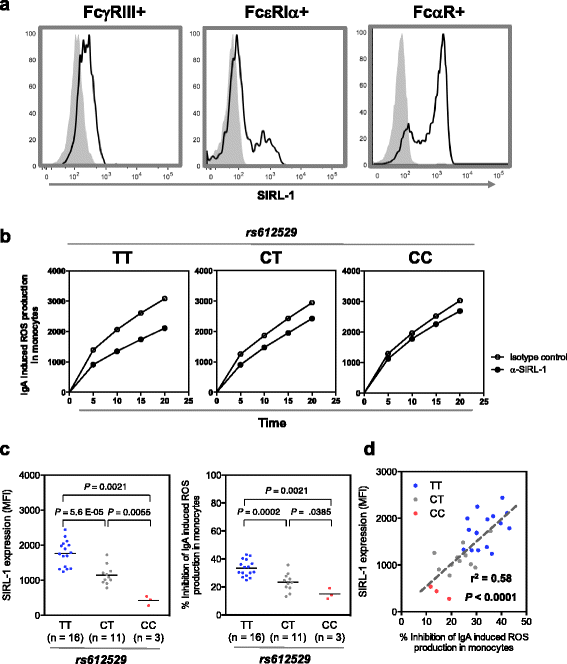
Results: We found a significant association of a single CpG-SNP, rs612529T/C, located in the promoter of VSTM1. Through flow cytometry analysis we confirmed that primarily in the monocytes the protein level of SIRL-1 is strongly associated with genotype of this SNP. In monocytes, the T allele of this SNP facilitates binding of the transcription factors YY1 and PU.1, of which the latter has been recently shown to act as docking site for modifiers of DNA methylation. In line with this notion rs612529T associates with a complete demethylation of the VSTM1 promoter correlating with the allele-specific upregulation of SIRL-1 expression. In monocytes, this upregulation strongly impacts the IgA-induced production of ROS by these cells. Through targeted association analysis we found a significant Meta P value of 1.14 × 10-6 for rs612529 for association to atopic dermatitis (AD).
Conclusion: Low expression of SIRL-1 on monocytes is associated with an increased risk for the manifestation of an inflammatory skin disease. It thus underlines the role of both the cell subset and this inhibitory immune receptor in maintaining immune homeostasis in the skin. Notably, the genetic regulation is achieved by a single CpG-SNP, which controls the overall methylation state of the promoter gene segment.
Keywords: Atopic dermatitis; Expression quantitative trait loci (eQTL); Monocytes; Neutrophils; Reactive oxygen species (ROS); Signal inhibitory receptor on leukocytes-1 (SIRL-1); VSTM1.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

