S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion
- PMID: 28220209
- PMCID: PMC5487892
- DOI: 10.1007/s00018-017-2474-4
S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion
Abstract
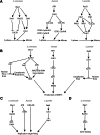
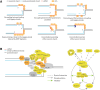
DNA replication stress, an important source of genomic instability, arises upon different types of DNA replication perturbations, including those that stall replication fork progression. Inhibitors of the cellular pool of deoxynucleotide triphosphates (dNTPs) slow down DNA synthesis throughout the genome. Following depletion of dNTPs, the highly conserved replication checkpoint kinase pathway, also known as the S-phase checkpoint, preserves the functionality and structure of stalled DNA replication forks and prevents chromosome fragmentation. The underlying mechanisms involve pathways extrinsic to replication forks, such as those involving regulation of the ribonucleotide reductase activity, the temporal program of origin firing, and cell cycle transitions. In addition, the S-phase checkpoint modulates the function of replisome components to promote replication integrity. This review summarizes the various functions of the replication checkpoint in promoting replication fork stability and genome integrity in the face of replication stress caused by dNTP depletion.
Keywords: ATR/Mec1/Rad3; Chromosome fragility; Fork remodeling; Helicases; Nucleases; Rad53/CHK1/Cds1; Stalled replication forks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases.Mol Cell. 2012 Mar 9;45(5):696-704. doi: 10.1016/j.molcel.2012.01.007. Epub 2012 Feb 9. Mol Cell. 2012. PMID: 22325992
-
The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes.Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2019183118. doi: 10.1073/pnas.2019183118. Proc Natl Acad Sci U S A. 2021. PMID: 34108240 Free PMC article.
-
The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability.Mol Cell. 2015 Sep 17;59(6):998-1010. doi: 10.1016/j.molcel.2015.07.030. Epub 2015 Sep 10. Mol Cell. 2015. PMID: 26365379 Free PMC article.
-
The Intra-S Checkpoint Responses to DNA Damage.Genes (Basel). 2017 Feb 17;8(2):74. doi: 10.3390/genes8020074. Genes (Basel). 2017. PMID: 28218681 Free PMC article. Review.
-
The S-phase checkpoint: targeting the replication fork.Biol Cell. 2009 Aug 19;101(11):617-27. doi: 10.1042/BC20090053. Biol Cell. 2009. PMID: 19686094 Review.
Cited by
-
Mechanisms of Hydroxyurea-Induced Cellular Senescence: An Oxidative Stress Connection?Oxid Med Cell Longev. 2021 Oct 18;2021:7753857. doi: 10.1155/2021/7753857. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34707779 Free PMC article. Review.
-
DNA damage responses that enhance resilience to replication stress.Cell Mol Life Sci. 2021 Nov;78(21-22):6763-6773. doi: 10.1007/s00018-021-03926-3. Epub 2021 Aug 31. Cell Mol Life Sci. 2021. PMID: 34463774 Free PMC article. Review.
-
The CDK-PLK1 axis targets the DNA damage checkpoint sensor protein RAD9 to promote cell proliferation and tolerance to genotoxic stress.Elife. 2017 Dec 19;6:e29953. doi: 10.7554/eLife.29953. Elife. 2017. PMID: 29254517 Free PMC article.
-
Rad53 arrests leading and lagging strand DNA synthesis via distinct mechanisms in response to DNA replication stress.Bioessays. 2022 Sep;44(9):e2200061. doi: 10.1002/bies.202200061. Epub 2022 Jul 1. Bioessays. 2022. PMID: 35778827 Free PMC article.
-
Rad9/53BP1 protects stalled replication forks from degradation in Mec1/ATR-defective cells.EMBO Rep. 2018 Feb;19(2):351-367. doi: 10.15252/embr.201744910. Epub 2018 Jan 4. EMBO Rep. 2018. PMID: 29301856 Free PMC article.
References
-
- Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

