Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects
- PMID: 28220799
- PMCID: PMC5321775
- DOI: 10.1038/ncomms14256
Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects
Abstract
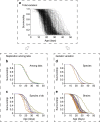
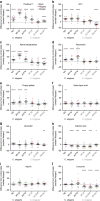
Limiting the debilitating consequences of ageing is a major medical challenge of our time. Robust pharmacological interventions that promote healthy ageing across diverse genetic backgrounds may engage conserved longevity pathways. Here we report results from the Caenorhabditis Intervention Testing Program in assessing longevity variation across 22 Caenorhabditis strains spanning 3 species, using multiple replicates collected across three independent laboratories. Reproducibility between test sites is high, whereas individual trial reproducibility is relatively low. Of ten pro-longevity chemicals tested, six significantly extend lifespan in at least one strain. Three reported dietary restriction mimetics are mainly effective across C. elegans strains, indicating species and strain-specific responses. In contrast, the amyloid dye ThioflavinT is both potent and robust across the strains. Our results highlight promising pharmacological leads and demonstrate the importance of assessing lifespans of discrete cohorts across repeat studies to capture biological variation in the search for reproducible ageing interventions.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

