Human enterovirus 71 protein interaction network prompts antiviral drug repositioning
- PMID: 28220872
- PMCID: PMC5318855
- DOI: 10.1038/srep43143
Human enterovirus 71 protein interaction network prompts antiviral drug repositioning
Abstract
As a predominant cause of human hand, foot, and mouth disease, enterovirus 71 (EV71) infection may lead to serious diseases and result in severe consequences that threaten public health and cause widespread panic. Although the systematic identification of physical interactions between viral proteins and host proteins provides initial information for the recognition of the cellular mechanism involved in viral infection and the development of new therapies, EV71-host protein interactions have not been explored. Here, we identified interactions between EV71 proteins and host cellular proteins and confirmed the functional relationships of EV71-interacting proteins (EIPs) with virus proliferation and infection by integrating a human protein interaction network and by functional annotation. We found that most EIPs had known interactions with other viruses. We also predicted ATP6V0C as a broad-spectrum essential host factor and validated its essentiality for EV71 infection in vitro. EIPs and their interacting proteins were more likely to be targets of anti-inflammatory and neurological drugs, indicating their potential to serve as host-oriented antiviral targets. Thus, we used a connectivity map to find drugs that inhibited EIP expression. We predicted tanespimycin as a candidate and demonstrated its antiviral efficiency in vitro. These findings provide the first systematic identification of EV71-host protein interactions, an analysis of EIP protein characteristics and a demonstration of their value in developing host-oriented antiviral therapies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
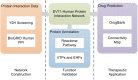



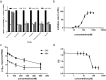
Similar articles
-
SUMO Modification Stabilizes Enterovirus 71 Polymerase 3D To Facilitate Viral Replication.J Virol. 2016 Nov 14;90(23):10472-10485. doi: 10.1128/JVI.01756-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27630238 Free PMC article.
-
Antiviral screen identifies EV71 inhibitors and reveals camptothecin-target, DNA topoisomerase 1 as a novel EV71 host factor.Antiviral Res. 2017 Jul;143:122-133. doi: 10.1016/j.antiviral.2017.04.008. Epub 2017 Apr 17. Antiviral Res. 2017. PMID: 28427827
-
Pyrrolidine dithiocarbamate inhibits enterovirus 71 replication by down-regulating ubiquitin-proteasome system.Virus Res. 2015 Jan 2;195:207-16. doi: 10.1016/j.virusres.2014.10.012. Epub 2014 Oct 23. Virus Res. 2015. PMID: 25456405
-
Development of antiviral agents toward enterovirus 71 infection.J Microbiol Immunol Infect. 2015 Feb;48(1):1-8. doi: 10.1016/j.jmii.2013.11.011. Epub 2014 Feb 21. J Microbiol Immunol Infect. 2015. PMID: 24560700 Review.
-
[Molecular Mechanism of Action of hnRNP K and RTN3 in the Replication of Enterovirus 71].Bing Du Xue Bao. 2015 Mar;31(2):197-200. Bing Du Xue Bao. 2015. PMID: 26164948 Review. Chinese.
Cited by
-
Identification and Analysis of Biomarkers Associated with Lipophagy and Therapeutic Agents for COVID-19.Viruses. 2024 Jun 7;16(6):923. doi: 10.3390/v16060923. Viruses. 2024. PMID: 38932215 Free PMC article.
-
Homologous recombination shapes the genetic diversity of African swine fever viruses.Vet Microbiol. 2019 Sep;236:108380. doi: 10.1016/j.vetmic.2019.08.003. Epub 2019 Aug 10. Vet Microbiol. 2019. PMID: 31500735 Free PMC article.
-
Shared molecular signatures between coronavirus infection and neurodegenerative diseases provide targets for broad-spectrum drug development.Sci Rep. 2023 Apr 4;13(1):5457. doi: 10.1038/s41598-023-29778-4. Sci Rep. 2023. PMID: 37015947 Free PMC article.
-
Prediction of antiviral drugs against African swine fever viruses based on protein-protein interaction analysis.PeerJ. 2020 Apr 1;8:e8855. doi: 10.7717/peerj.8855. eCollection 2020. PeerJ. 2020. PMID: 32274268 Free PMC article.
-
The RNA-dependent RNA polymerase of enterovirus A71 associates with ribosomal proteins and positively regulates protein translation.RNA Biol. 2020 Apr;17(4):608-622. doi: 10.1080/15476286.2020.1722448. Epub 2020 Feb 6. RNA Biol. 2020. PMID: 32009553 Free PMC article.
References
-
- Solomon T. et al.. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10, 778–790 (2010). - PubMed
-
- Cardosa M. J., Krishnan S., Tio P. H., Perera D. & Wong S. C. Isolation of subgenus B adenovirus during a fatal outbreak of enterovirus 71-associated hand, foot, and mouth disease in Sibu, Sarawak. Lancet 354, 987–991 (1999). - PubMed
-
- Ho M. et al.. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341, 929–935 (1999). - PubMed
-
- Chan L. G. et al.. Deaths of Children during an Outbreak of Hand, Foot, and Mouth Disease in Sarawak, Malaysia: Clinical and Pathological Characteristics of the Disease. Clin. Infect. Dis. 31, 678–683 (2000). - PubMed
-
- Chong C. Y. et al.. Hand, foot and mouth disease in Singapore: a comparison of fatal and non-fatal cases. Acta Paediatr. Oslo Nor. 1992 92, 1163–1169 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

