Early Transcriptomic Changes in the Ileal Pouch Provide Insight into the Molecular Pathogenesis of Pouchitis and Ulcerative Colitis
- PMID: 28221248
- PMCID: PMC5988644
- DOI: 10.1097/MIB.0000000000001027
Early Transcriptomic Changes in the Ileal Pouch Provide Insight into the Molecular Pathogenesis of Pouchitis and Ulcerative Colitis
Abstract
Background: Ulcerative colitis (UC) only involves the colonic mucosa. Yet, nearly 50% of patients with UC who undergo total proctocolectomy with ileal pouch anal anastomosis develop UC-like inflammation of the ileal pouch (pouchitis). By contrast, patients with familial adenomatous polyposis (FAP) with ileal pouch anal anastomosis develop pouchitis far less frequently. We hypothesized that pathogenic events associated with the development of UC are recapitulated by colonic-metaplastic transcriptomic reprogramming of the UC pouch.
Methods: We prospectively sampled pouch and prepouch ileum mucosal biopsies in patients with UC with ileal pouch anal anastomosis 4, 8, and 12 months after their pouch was in continuity. Mucosal samples were also obtained from patients with FAP. Transcriptional profiles of the UC and FAP pouch and prepouch ileum were investigated via RNA sequencing and compared with data from a previously published microarray study.
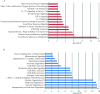
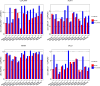
Results: Unlike patients with FAP, subjects with UC exhibited a large set of differentially expressed genes between the pouch and prepouch ileum as early as 4 months after pouch functionalization. Functional pathway analysis of differentially expressed genes in the UC pouch revealed an enhanced state of immune/inflammatory response and extracellular matrix remodeling. Moreover, >70% of differentially expressed genes mapped to published inflammatory bowel diseases microarray data sets displayed directional changes consistent with active UC but not with Crohn's disease.
Conclusions: The UC pouch, well before histologic inflammation, already displays a systems-level gain of colon-associated genes and loss of ileum-associated genes. Patients with UC exhibit a unique transcriptomic response to ileal pouch creation that can be observed well before disease and may in part explain their susceptibility to the development of pouchitis.
Figures




References
-
- Hurst RD, Molinari M, Chung TP, Rubin M, Michelassi F. Prospective study of the incidence, timing and treatment of pouchitis in 104 consecutive patients after restorative proctocolectomy. Arch Surg. 1996;131(5):492–497. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

