Circadian deep sequencing reveals stress-response genes that adopt robust rhythmic expression during aging
- PMID: 28221375
- PMCID: PMC5321795
- DOI: 10.1038/ncomms14529
Circadian deep sequencing reveals stress-response genes that adopt robust rhythmic expression during aging
Abstract
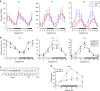
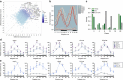
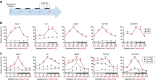
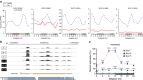
Disruption of the circadian clock, which directs rhythmic expression of numerous output genes, accelerates aging. To enquire how the circadian system protects aging organisms, here we compare circadian transcriptomes in heads of young and old Drosophila melanogaster. The core clock and most output genes remained robustly rhythmic in old flies, while others lost rhythmicity with age, resulting in constitutive over- or under-expression. Unexpectedly, we identify a subset of genes that adopted increased or de novo rhythmicity during aging, enriched for stress-response functions. These genes, termed late-life cyclers, were also rhythmically induced in young flies by constant exposure to exogenous oxidative stress, and this upregulation is CLOCK-dependent. We also identify age-onset rhythmicity in several putative primary piRNA transcripts overlapping antisense transposons. Our results suggest that, as organisms age, the circadian system shifts greater regulatory priority to the mitigation of accumulating cellular stress.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

